1. Home
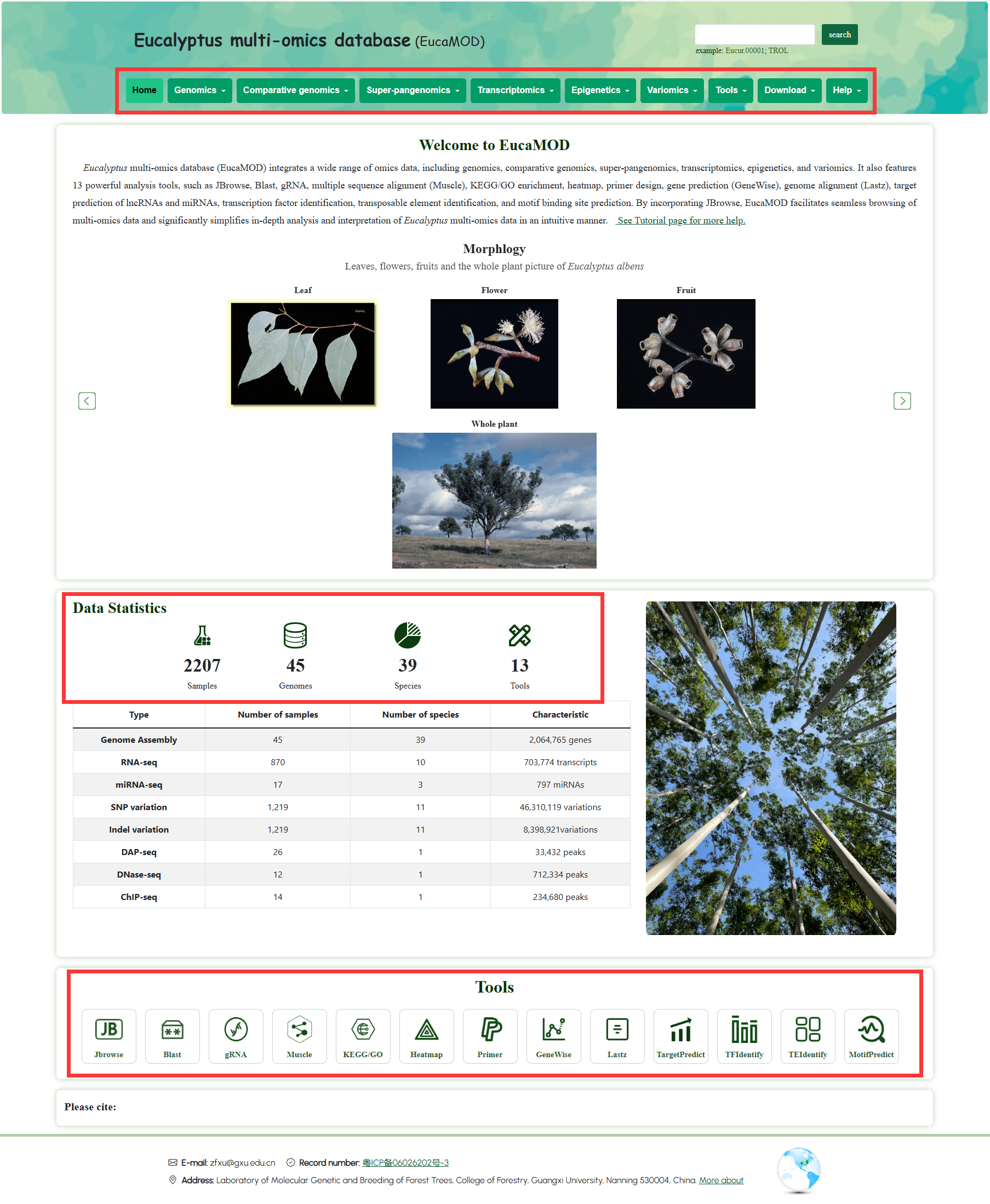
The homepage of EucaMOD displays the general navigation bar, basic information, quick tools, and contact details. EucaMOD provides 10 main functional modules: Home, Genomics, Comparative genomics, Pan-proteomics, Transcriptomics, Epigenetics, Variomics, Tools, Download, and Help. After inputting the gene ID/name/symbol of interest, you can quickly search the entire database and obtain multi-omics information through a one-click access on the homepage.
2. Genomics
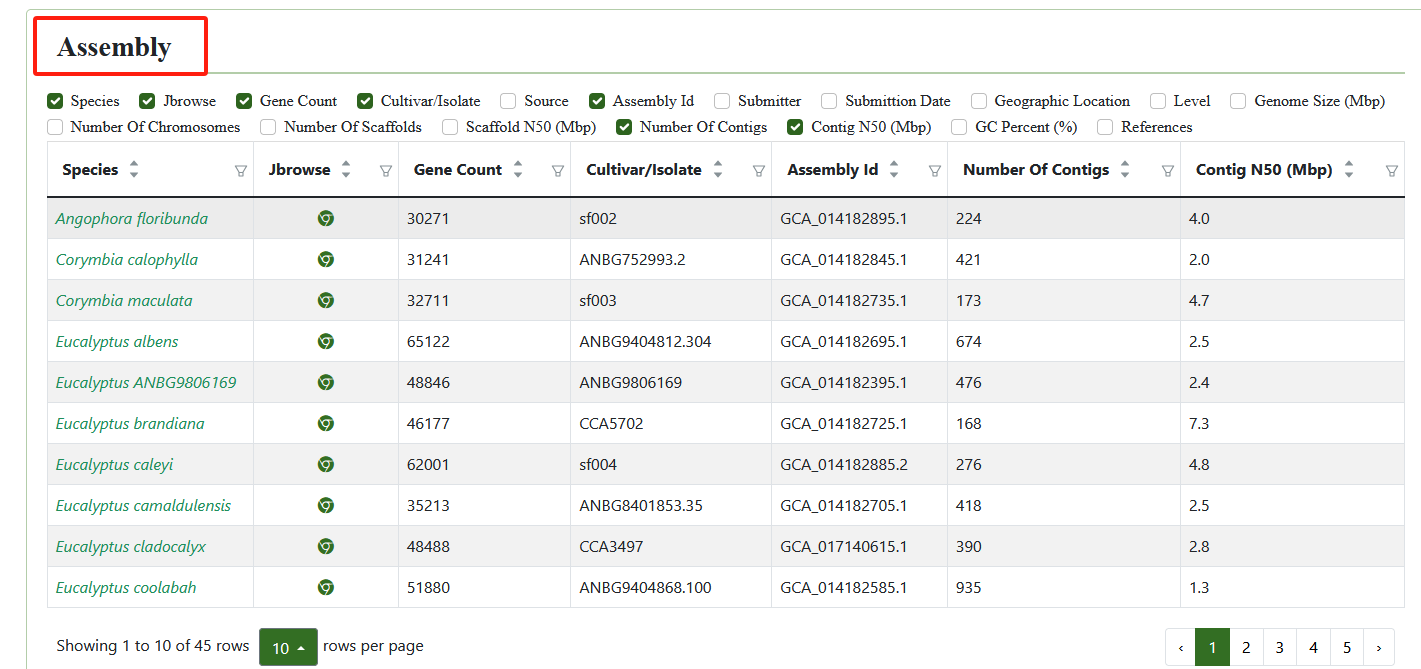
2.1. Assembly
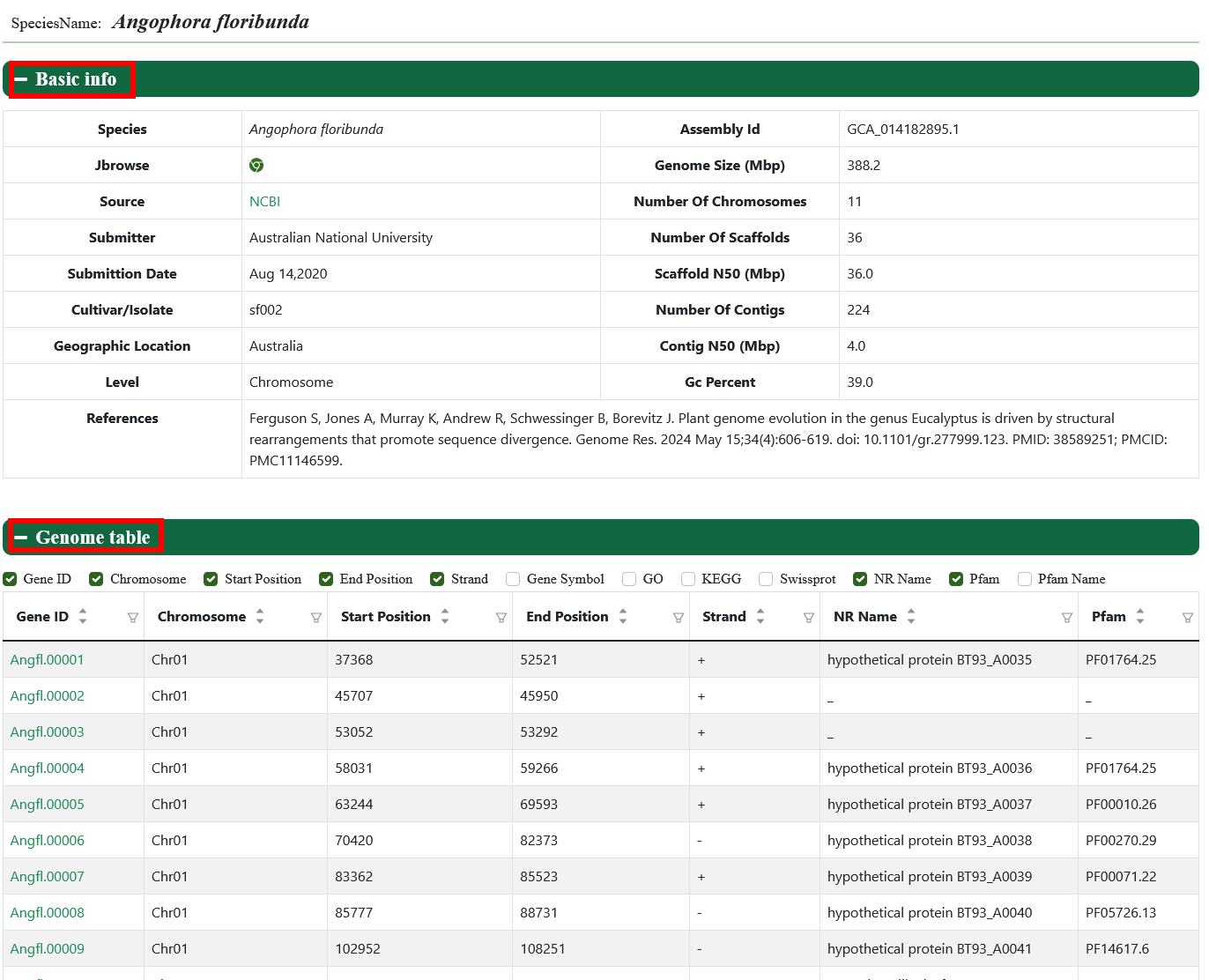
The genomics module collects 45 published genome assemblies. Users can click on the genome ID to access detailed information about the genome, such as its data source, total sequence length, chromosome length, gene information and etc.
2.2. Gene search
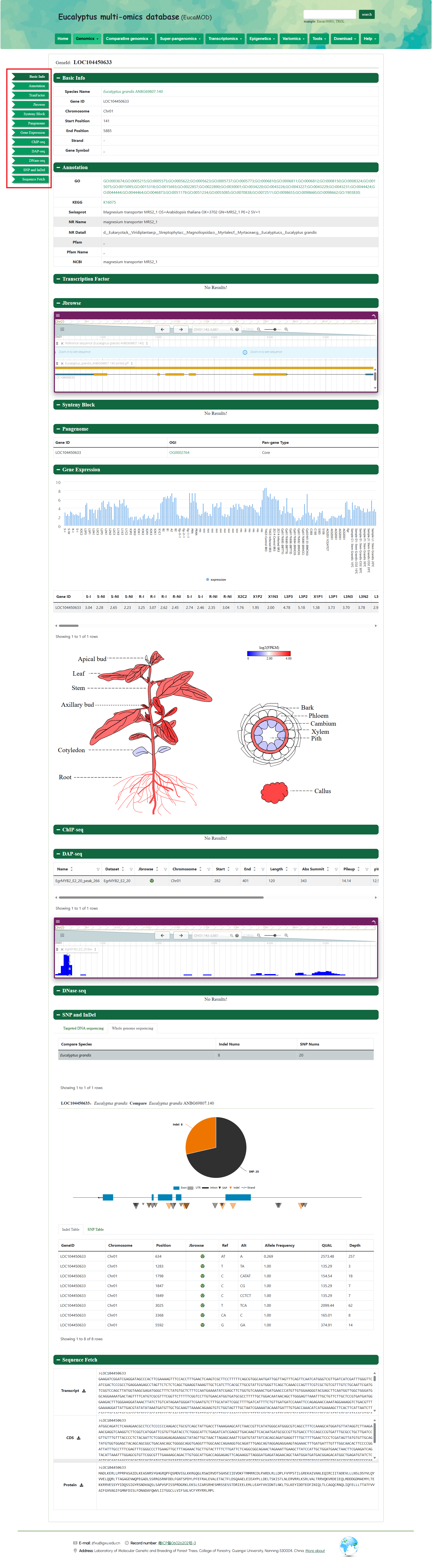
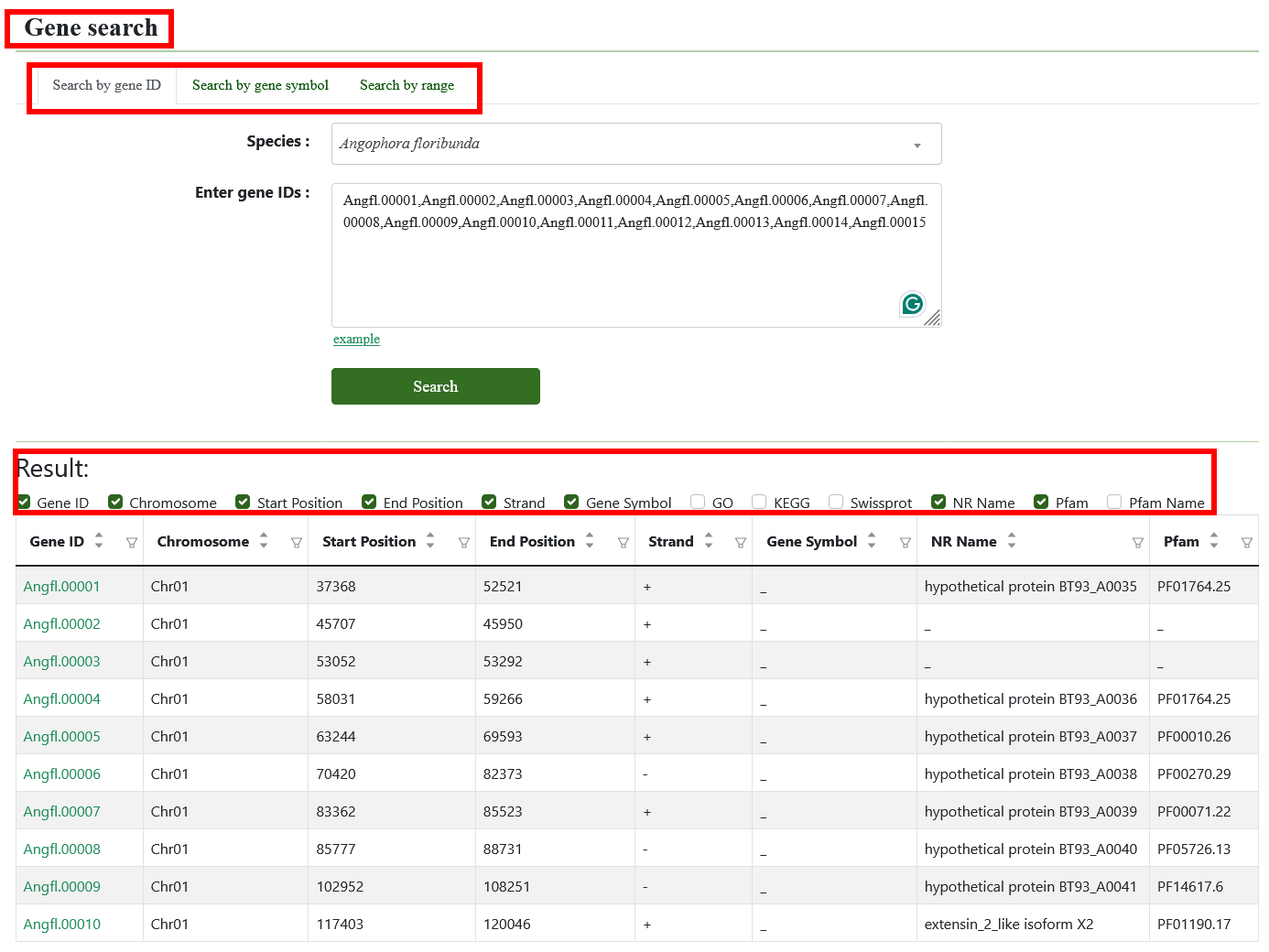
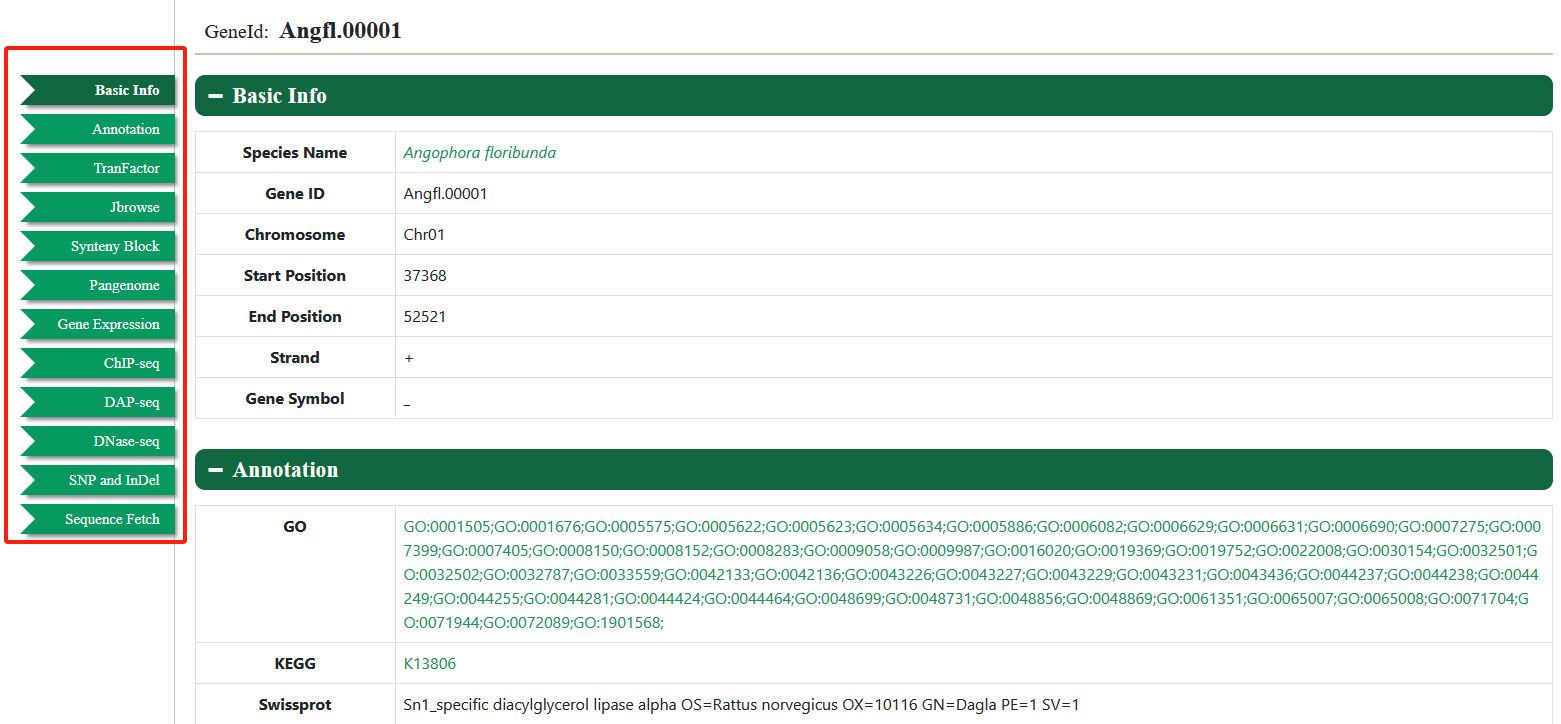
The gene search page provides gene structure and function information of Eucalyptus genomes. Various query modes are provided in this module. The users can enter the gene ID, gene name, genomic region, and gene list of interest, to obtain the structure and function information. Gene IDs can link to multiple omics datasets and can be viewed in JBrowse.
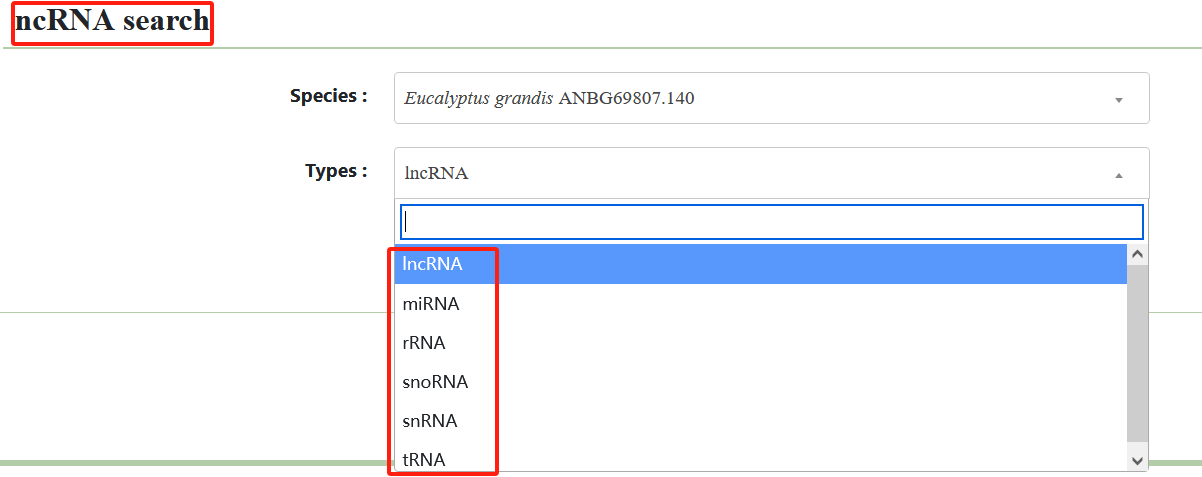
2.3. ncRNA search
We also provide non-coding RNA search.
2.4. Sequence fetch
Various query modes are provided in this module. The users can enter the gene ID, gene symbol, genomic region, and gene list of interest, to get sequences that you are interested.
3. Comparative genomics
3.1. Phylogenetic tree
Based on the single-copy gene family, we constructed a phylogenetic tree to show the evolution of different Eucalyptus species. We can view pictures of different Eucalyptus species on this page, including leaf, flower, fruit and whole plant.


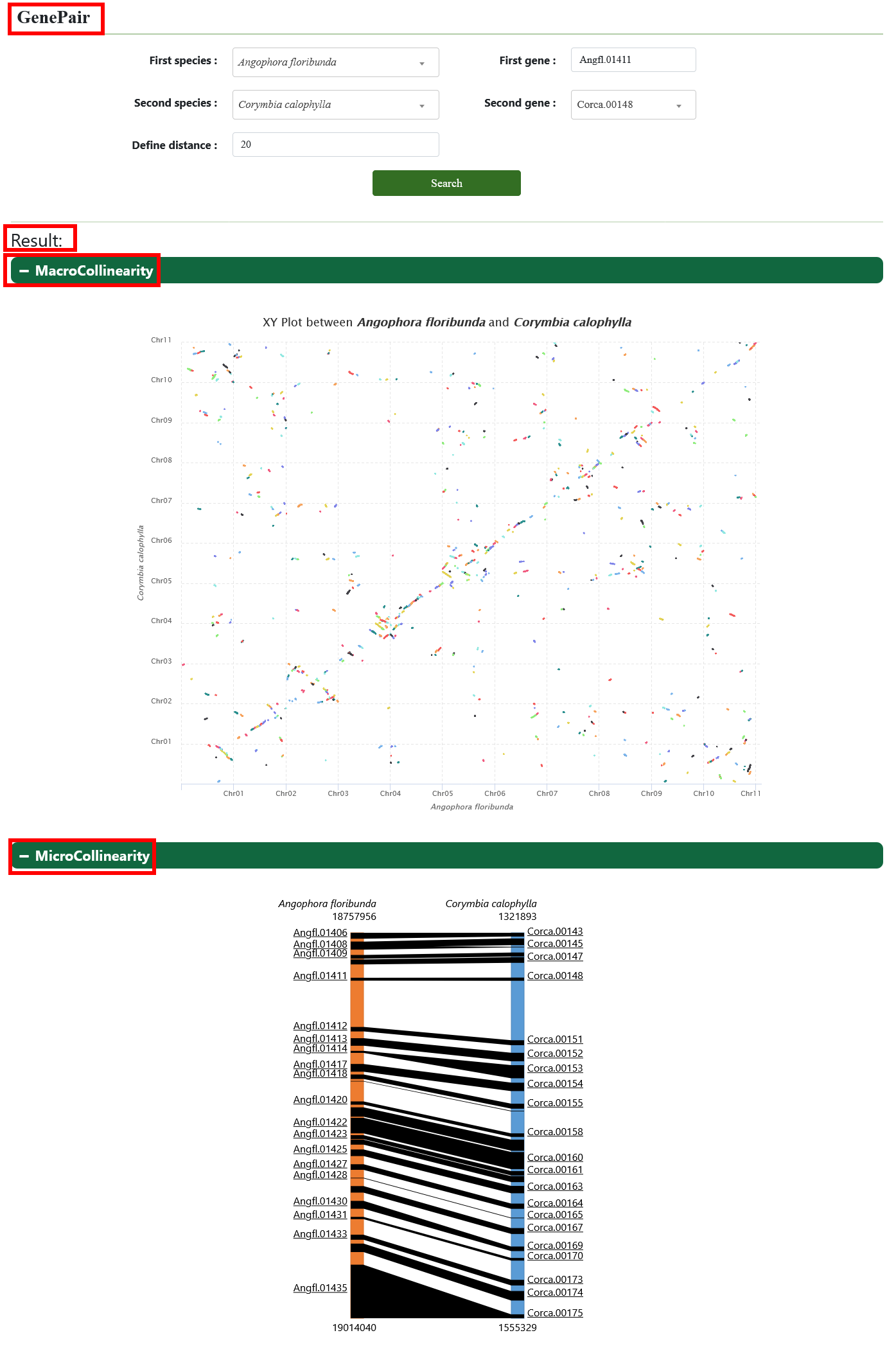
3.2. GenePair
The GenePair tool is used to query orthologous gene collinearity at gene levels.
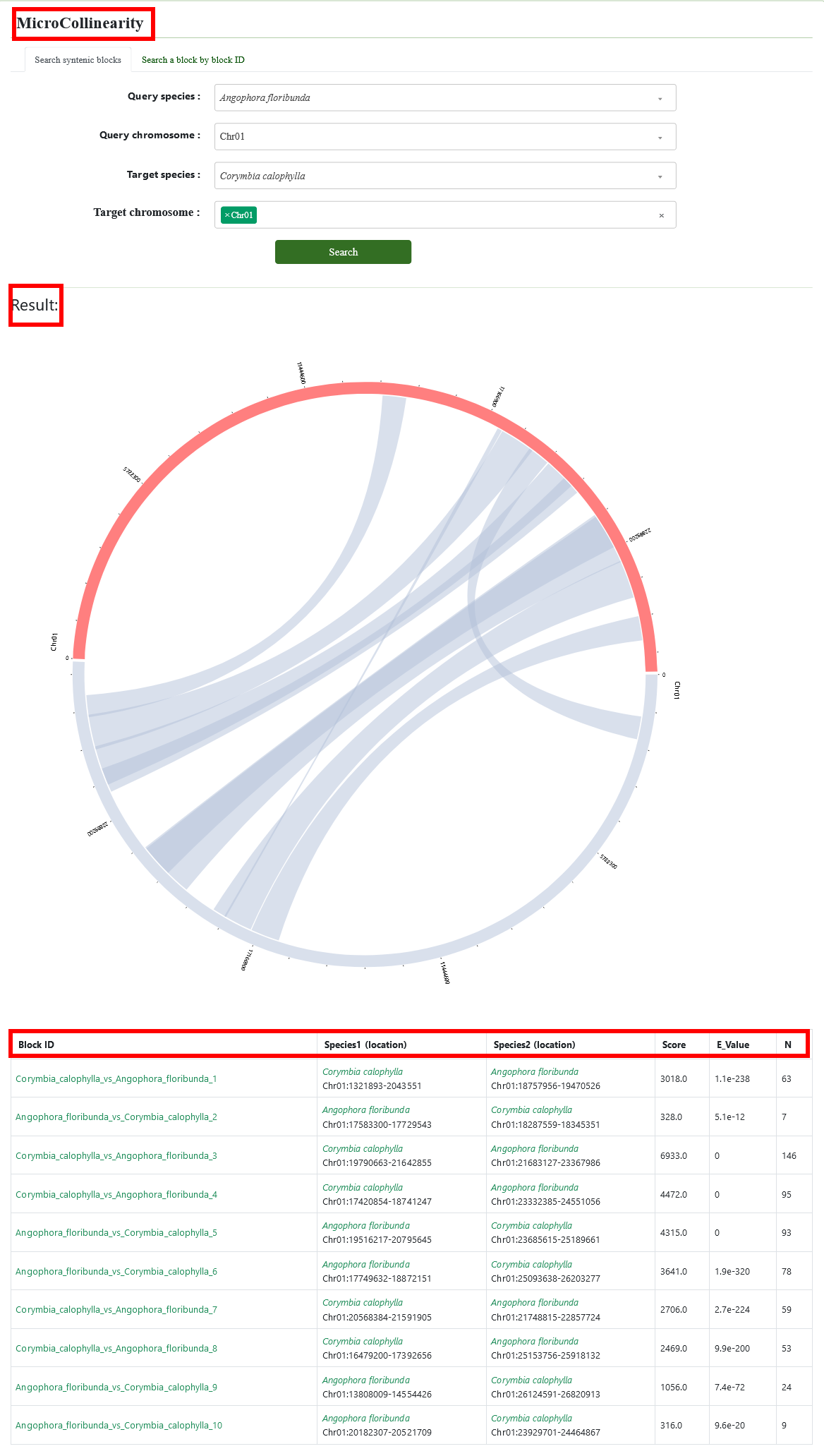
3.3. MicroCollinearity
The MicroCollinearity tool is used to query orthologous gene collinearity at chromosome levels.
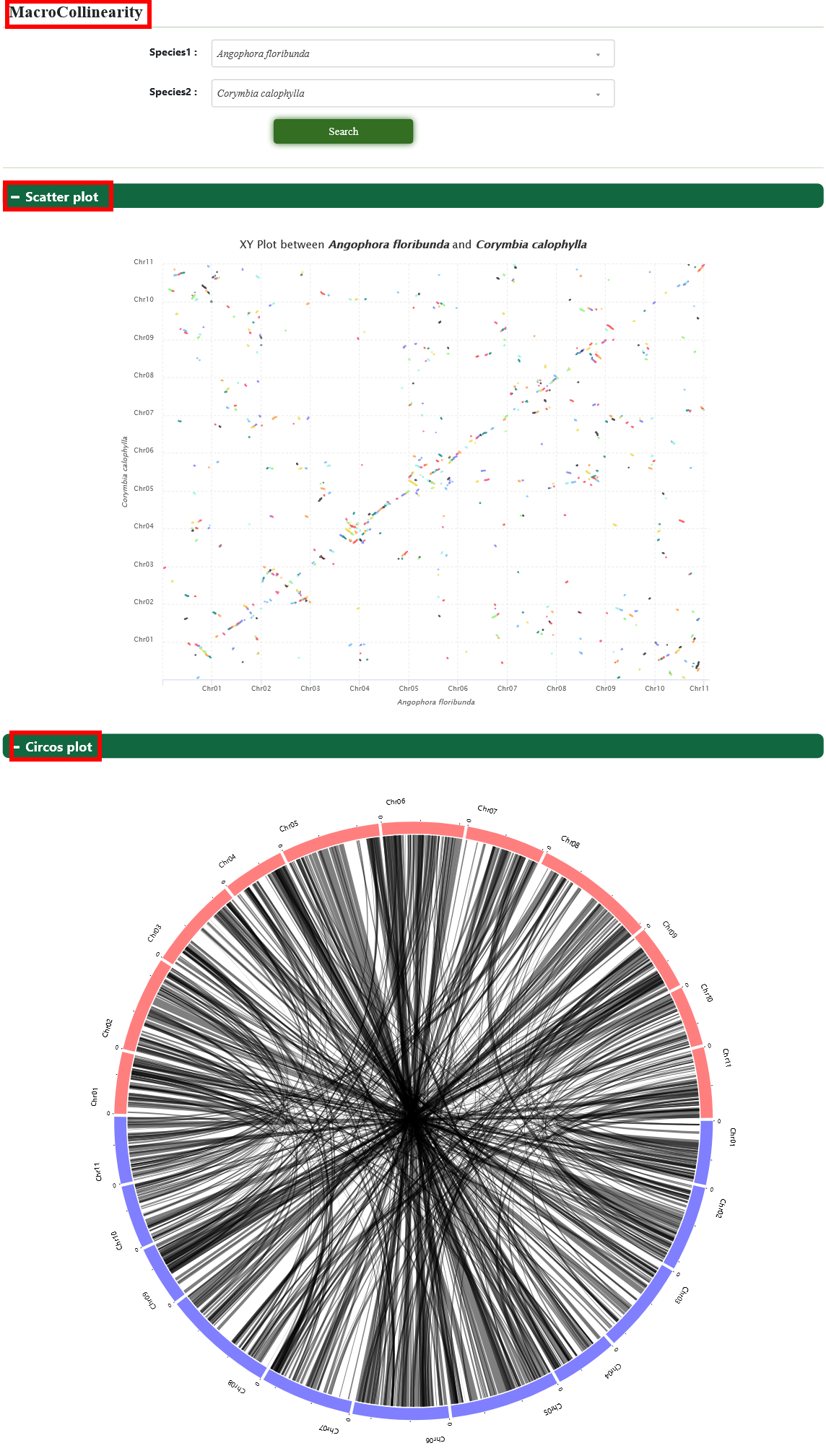
3.4. MacroCollinearity
The MacroCollinearity tool is constructed to the comparative genomic analysis of genome synteny.
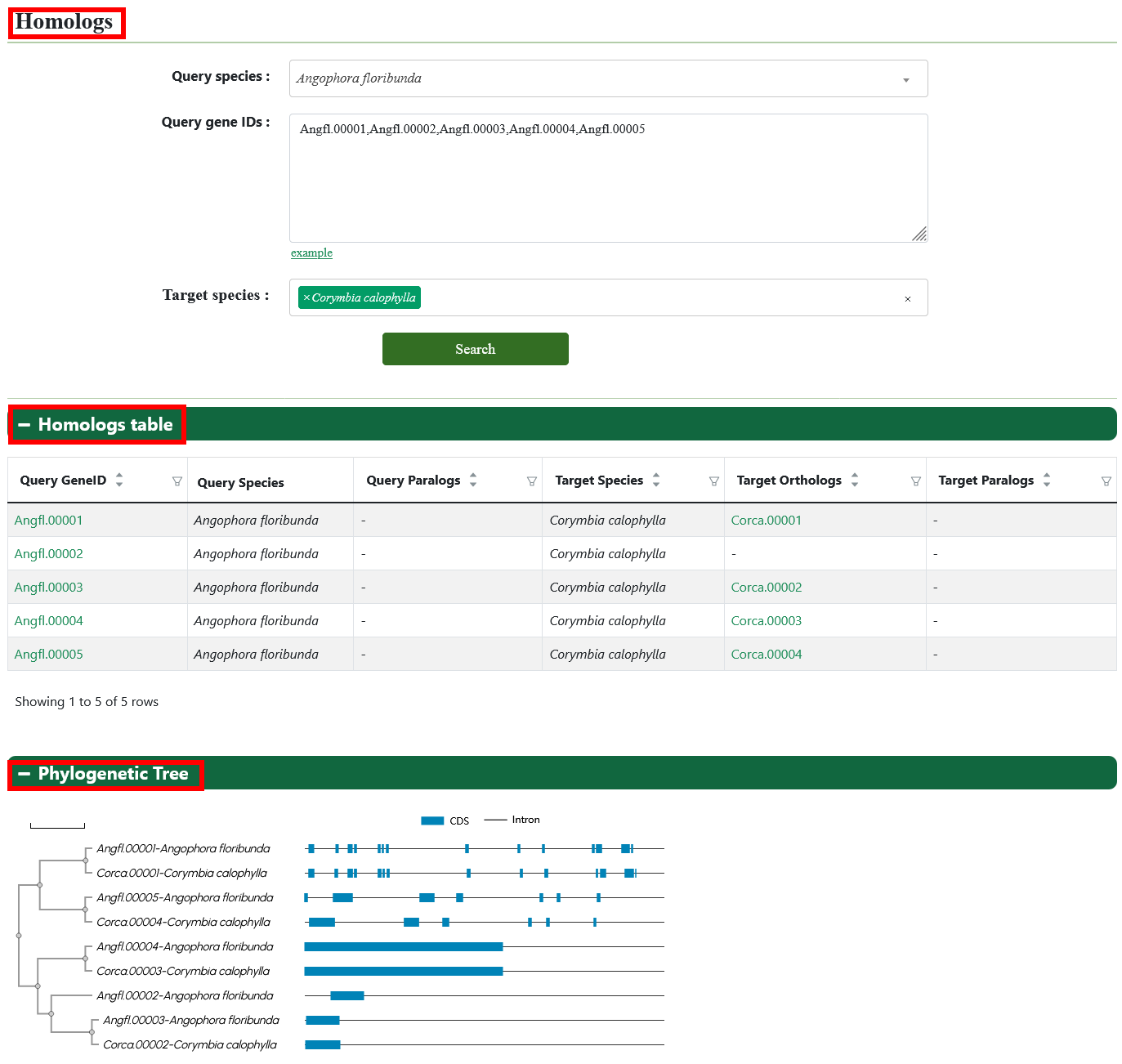
3.5. Homologs
The users can enter the query genome, gene ID, and target genome to obtain all homologous genes and their phylogenetic tree.
4. Super-pan-proteome
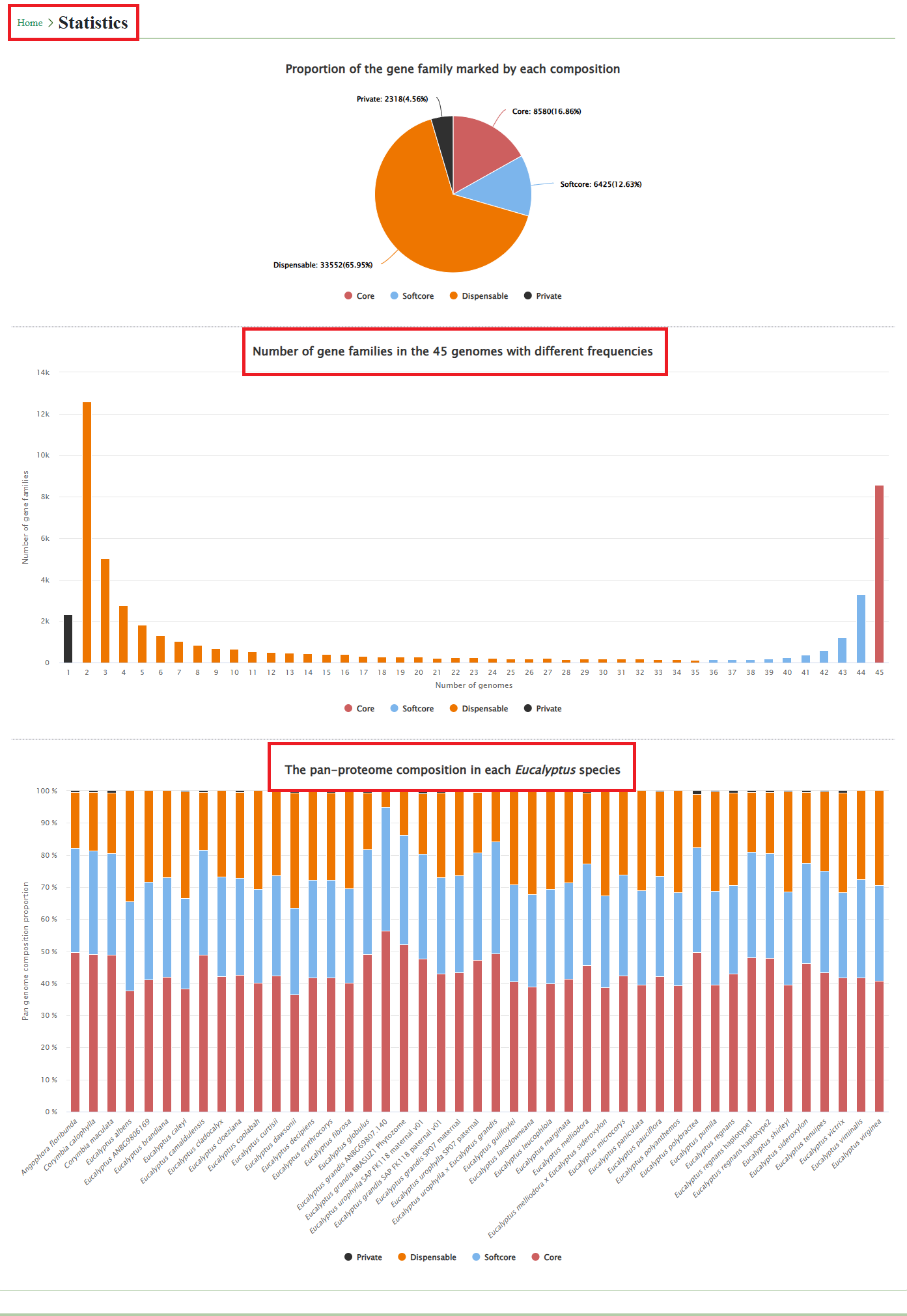
4.1. Statistic
The number of different gene families for each Eucalyptus species is also shown on the Statistic page.
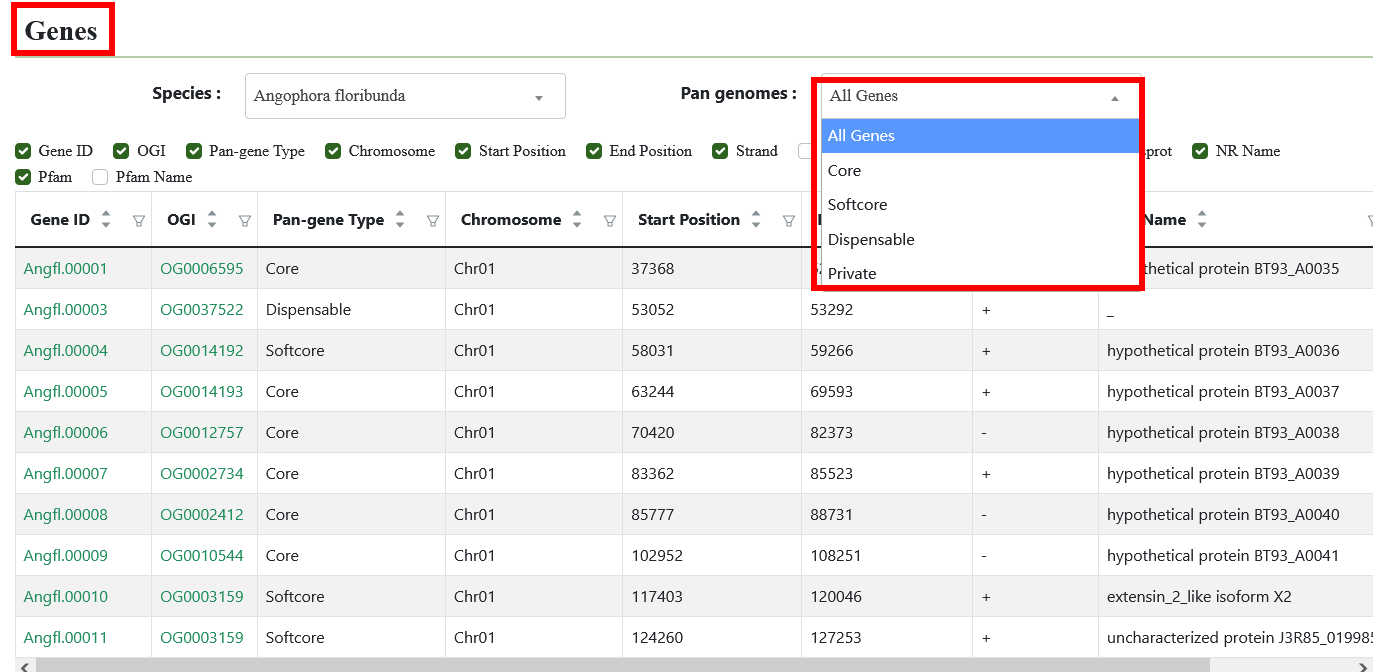
4.2. Genes
Users can query the core, softcore, dispensable, and species-specific genes of each Eucalyptus species in pangenome through this interface.
5. Transcriptomics
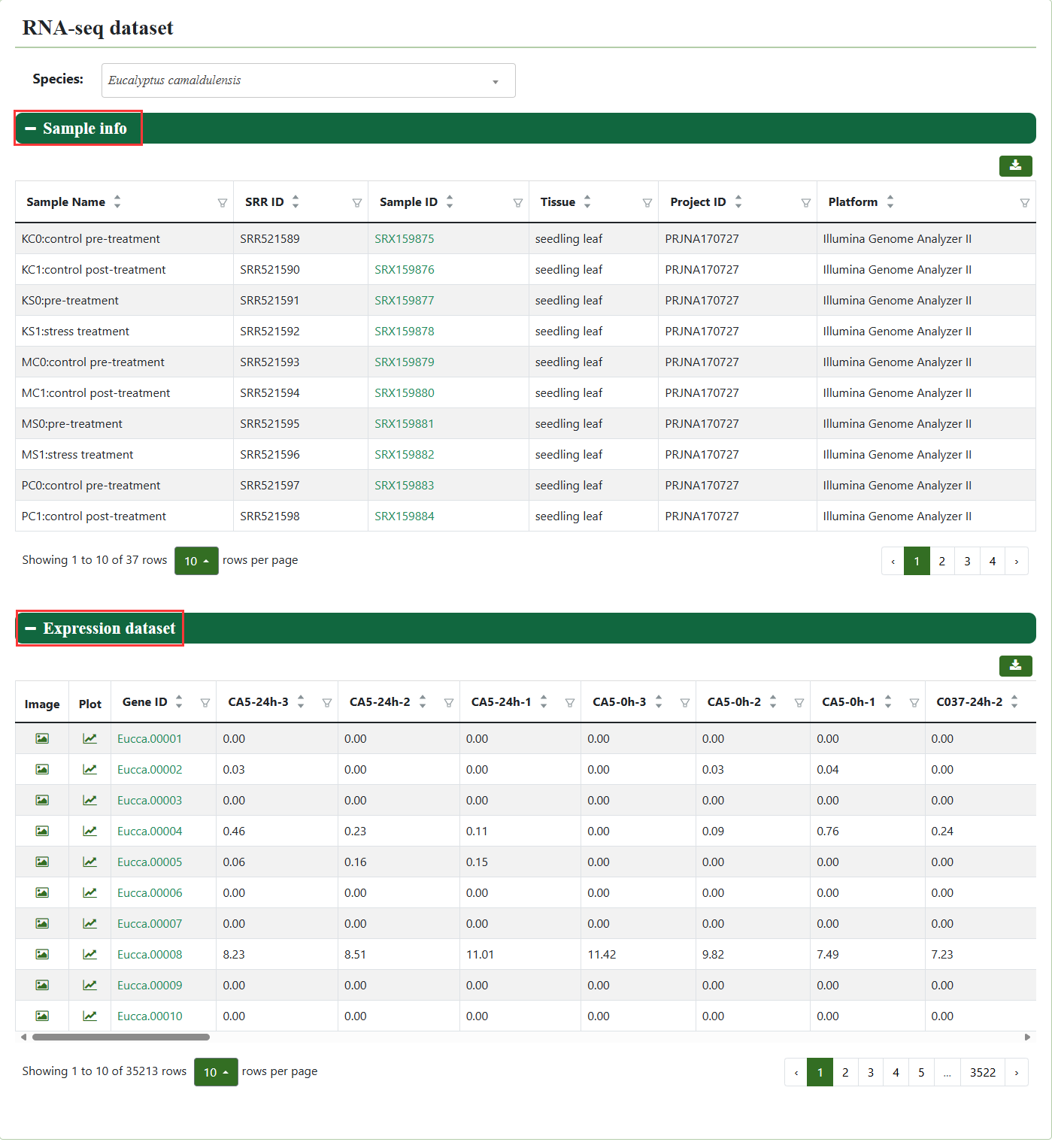
5.1. RNA-seq dataset
In the RNA-seq dataset, users can view sample information and the expression of genes of interest in all samples.
5.2. Gene expression profile
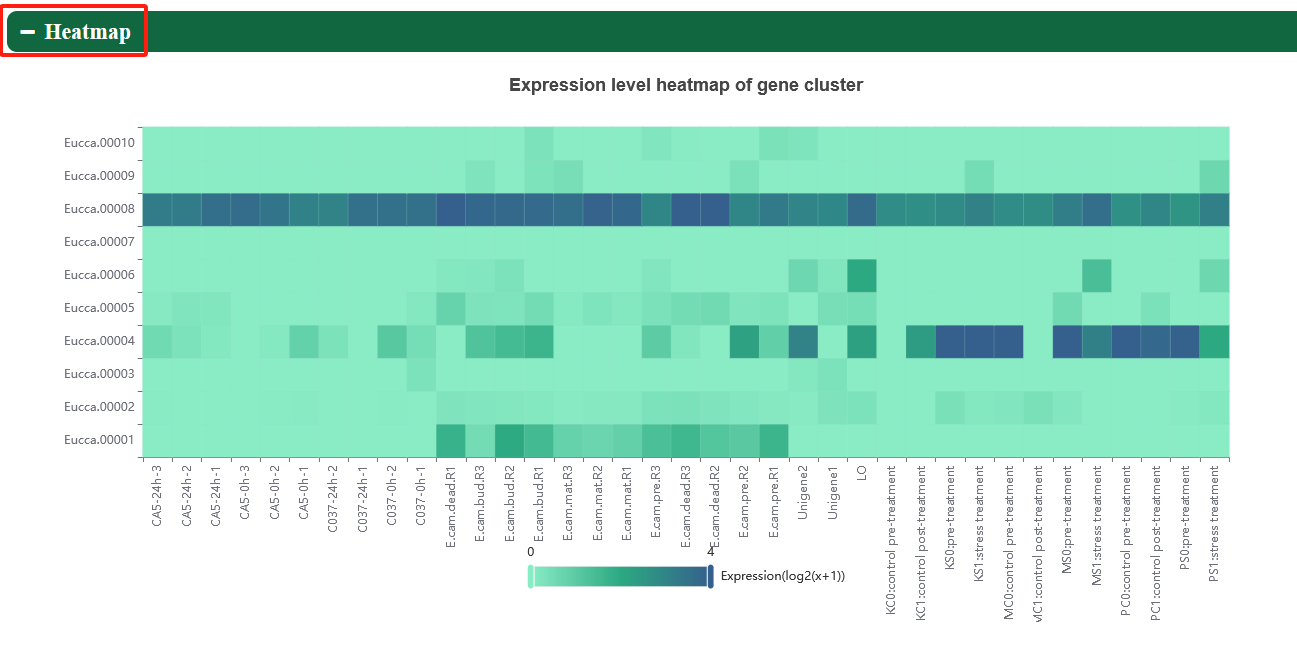
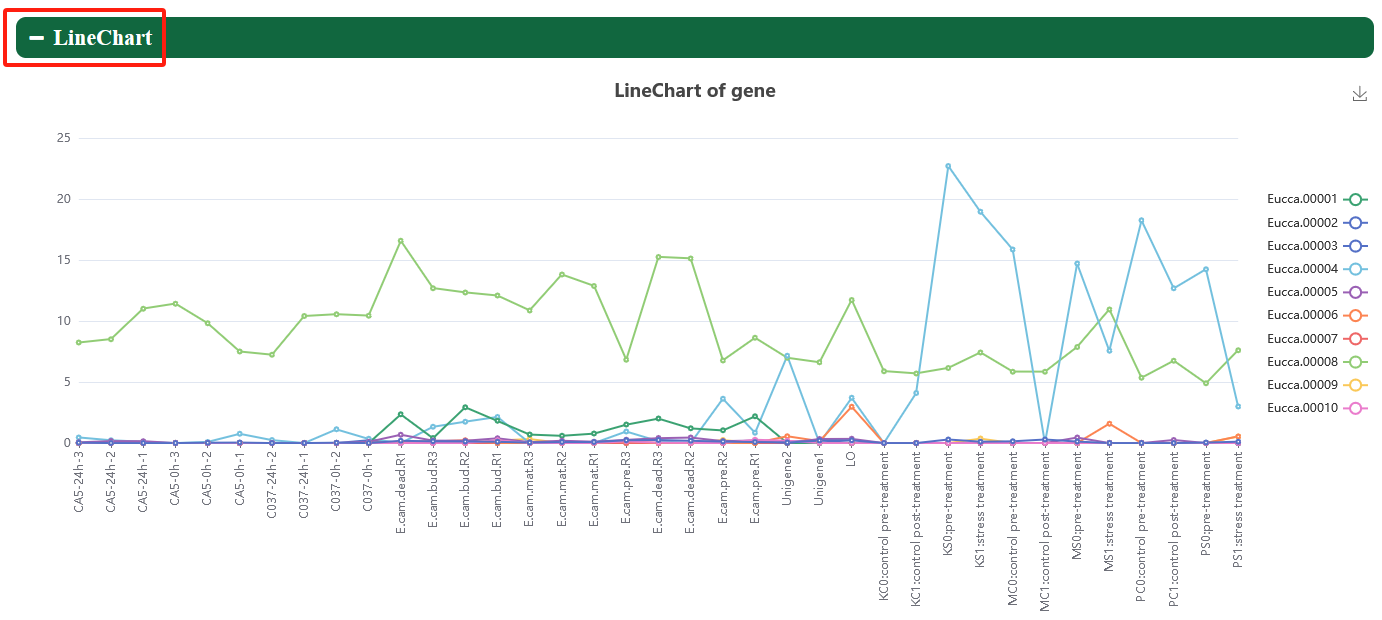
In order to meet specific research interests, we have added category labels and support eFP, heatmap, linechart, lineplot, and boxplot to visualize expression profiles of genes in different tissues.
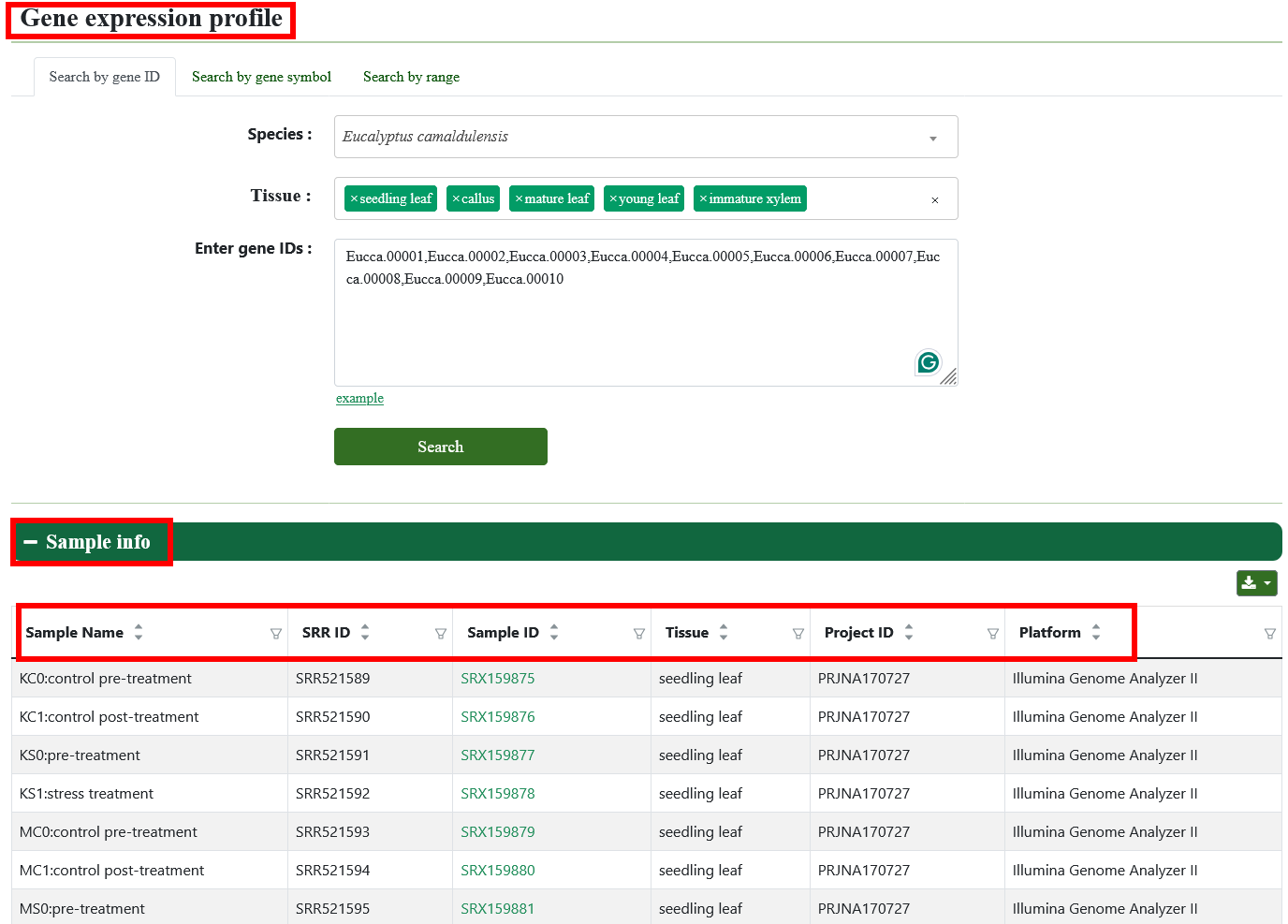
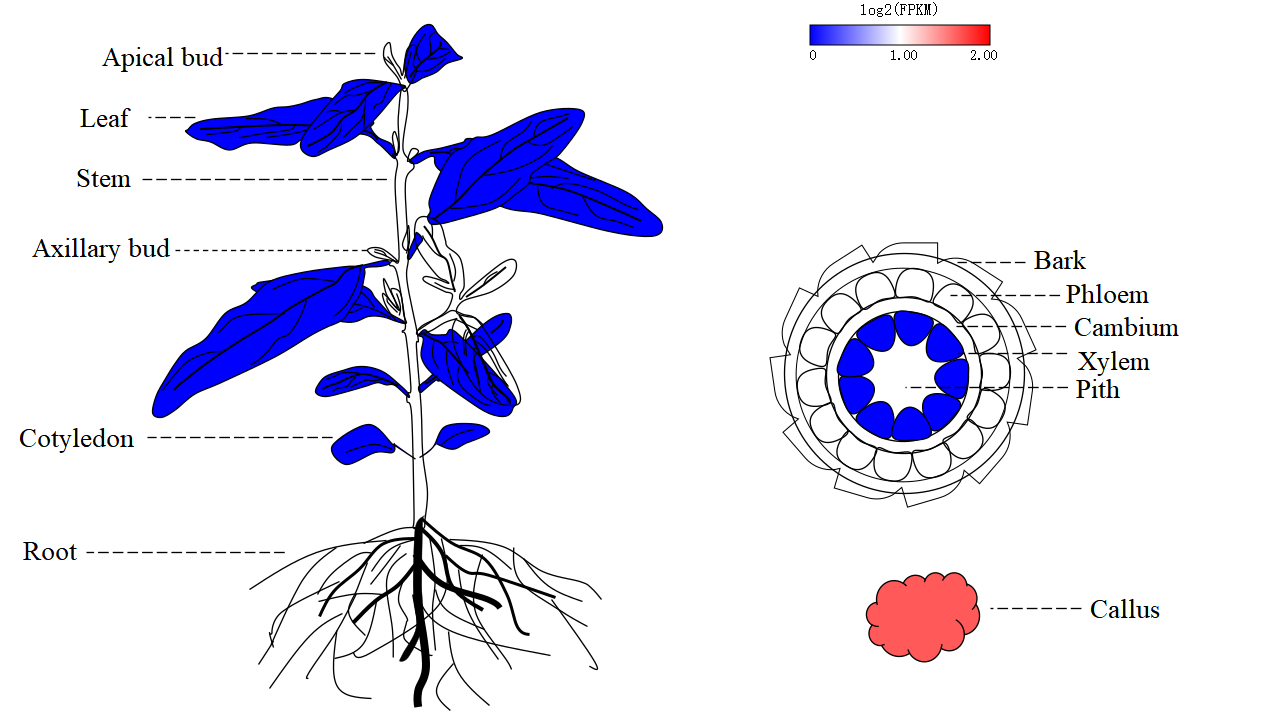
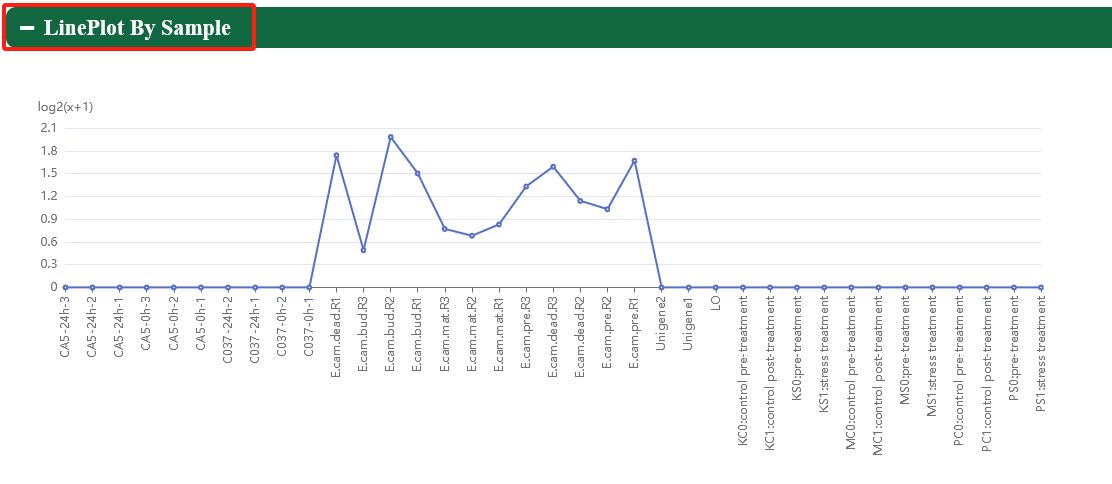
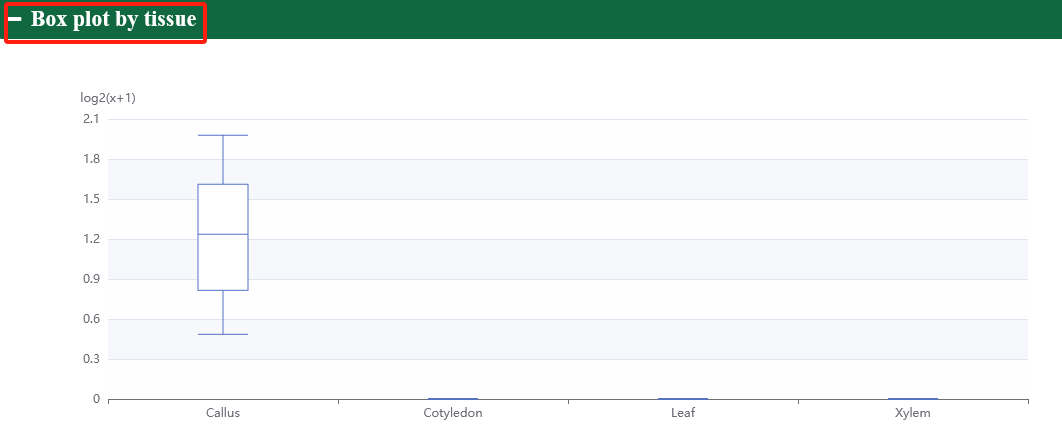
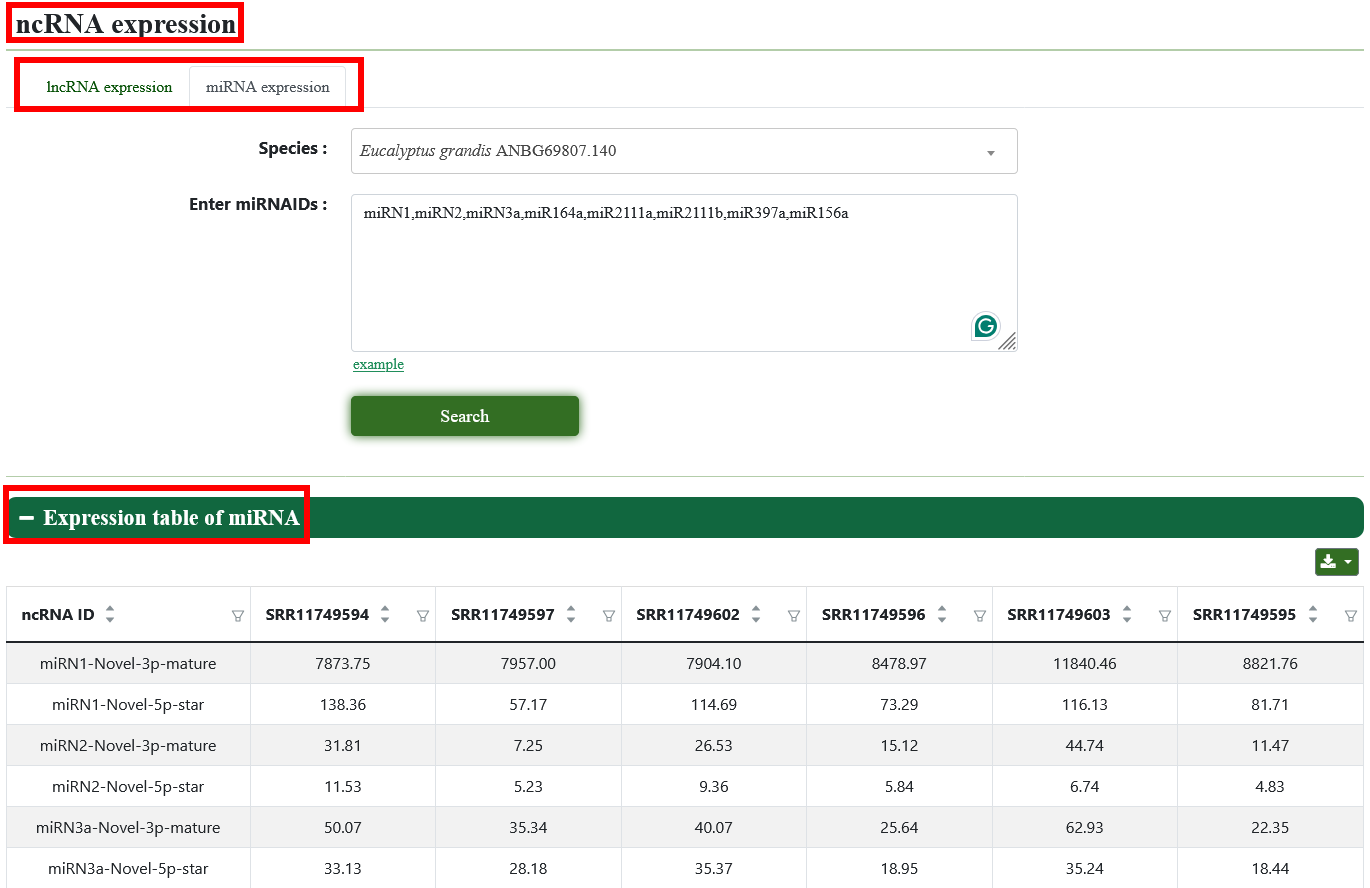
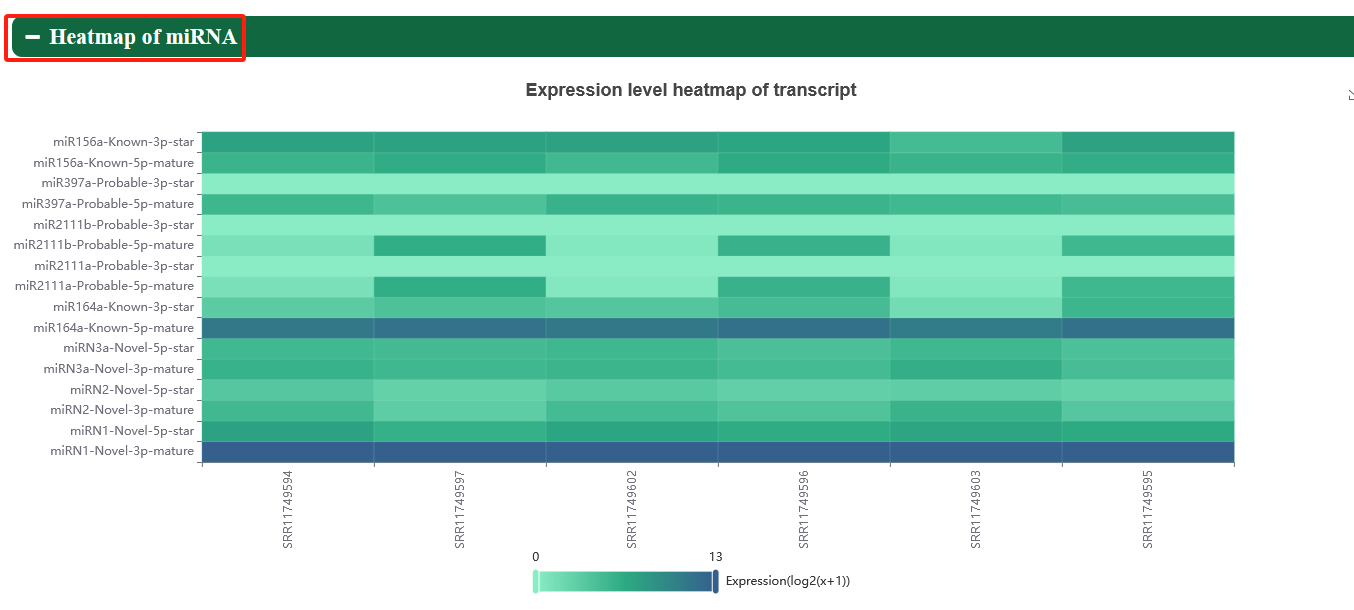
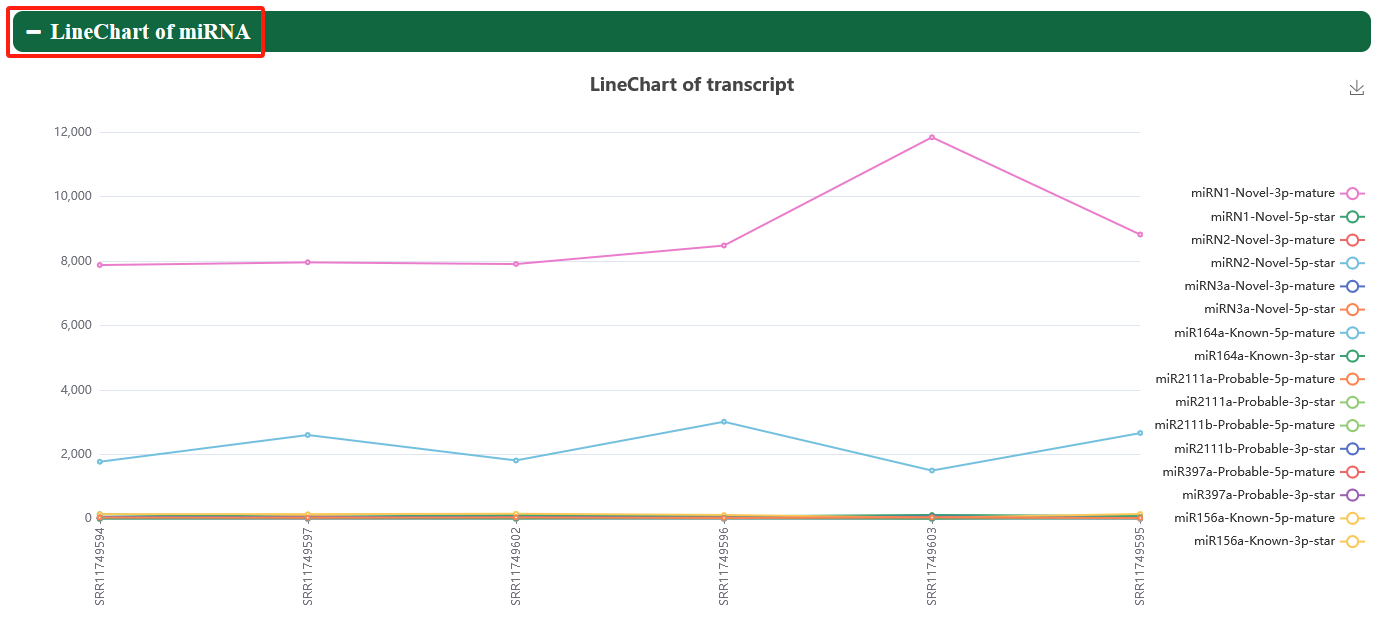
5.3. ncRNA expression
Users can view the lncRNA and miRNA of interest in samples of different Eucalyptus species.
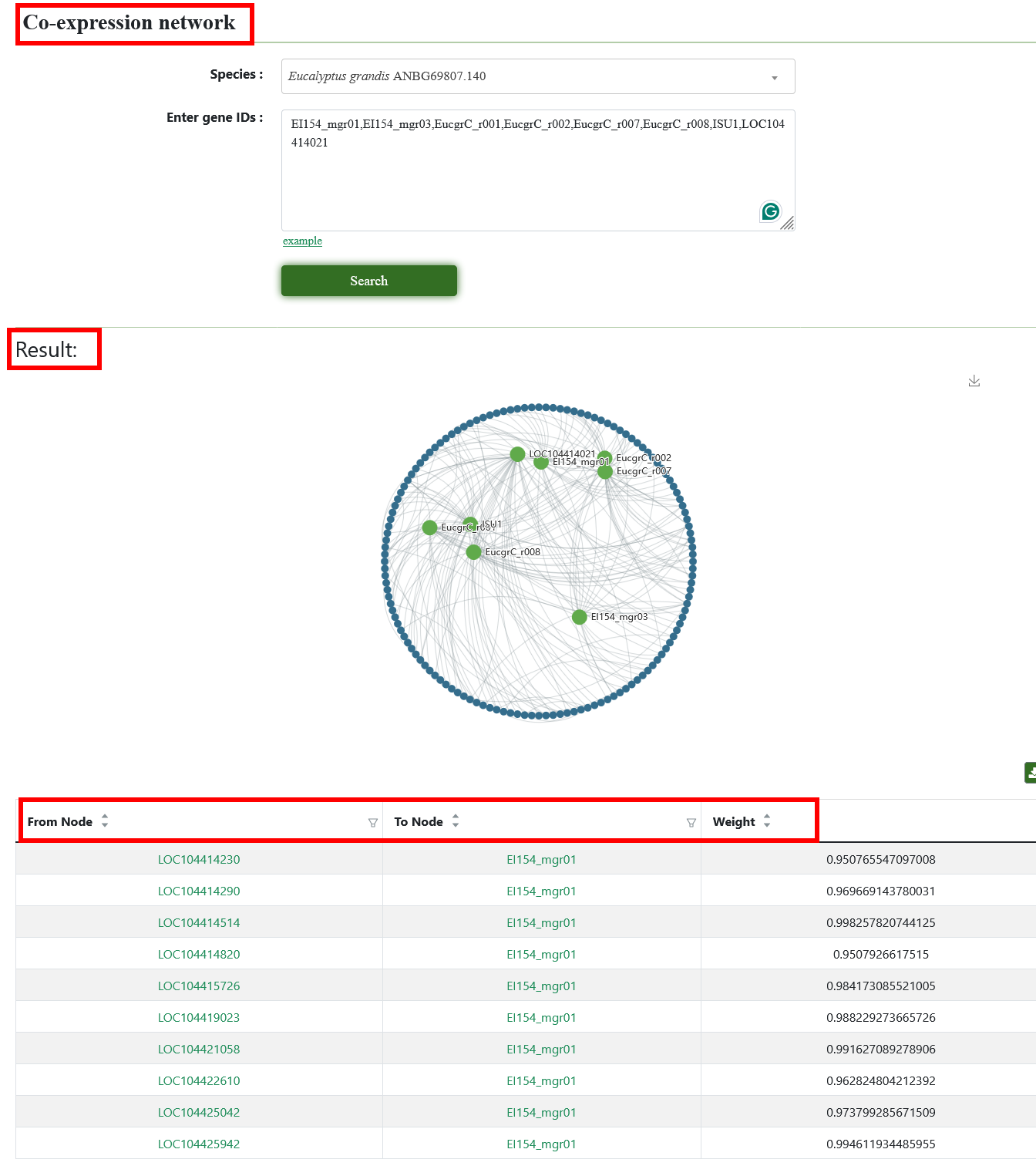
5.4. Co-expression network
Users need to select genome and enter Gene ID or list. After submission, the result is a network which shows the interaction between input genes.
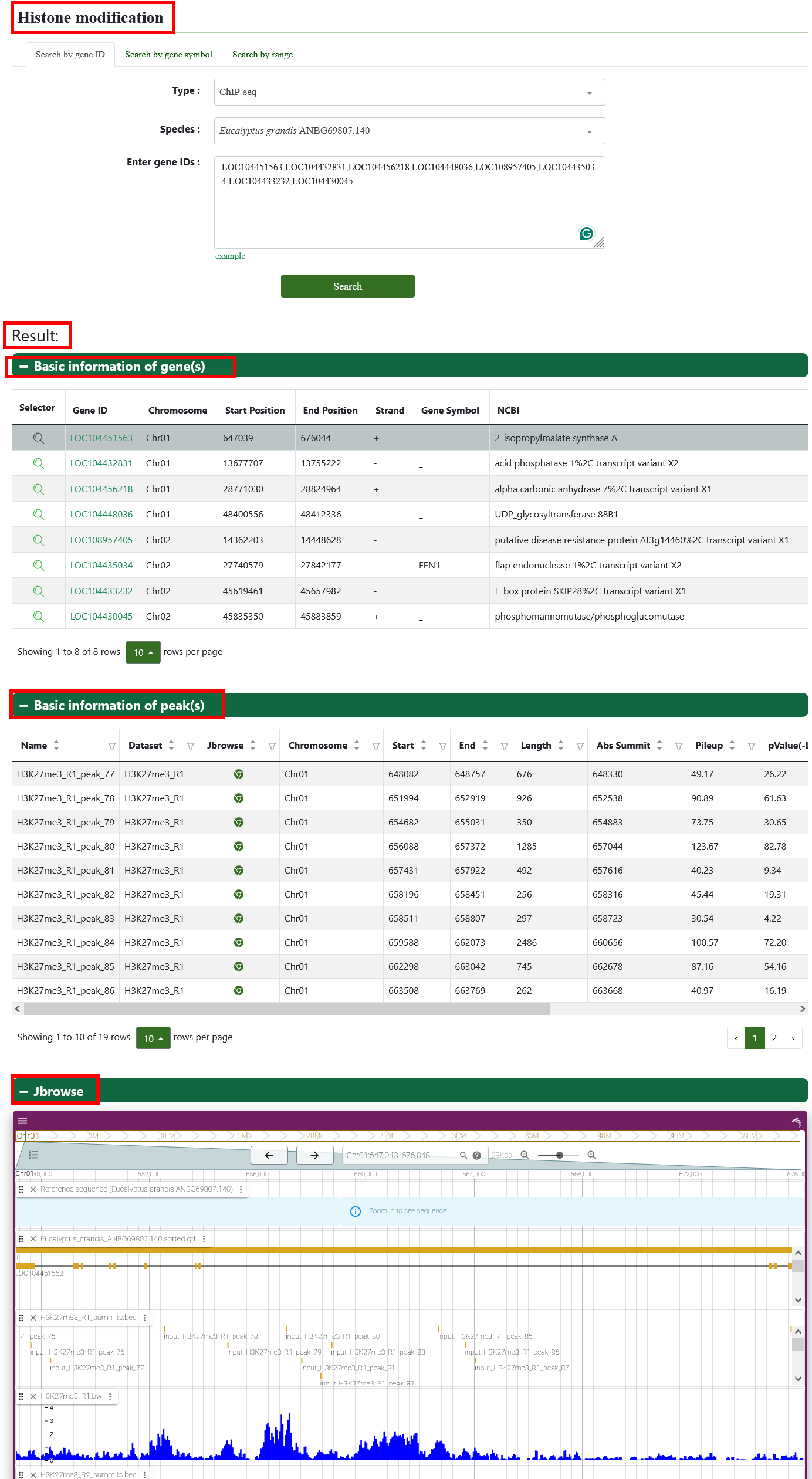
6. Epigenomics
6.1. Histone modification
We provide a visual view of histone modification in Eucalyptus grandis on the page and click to Jbrowse page.
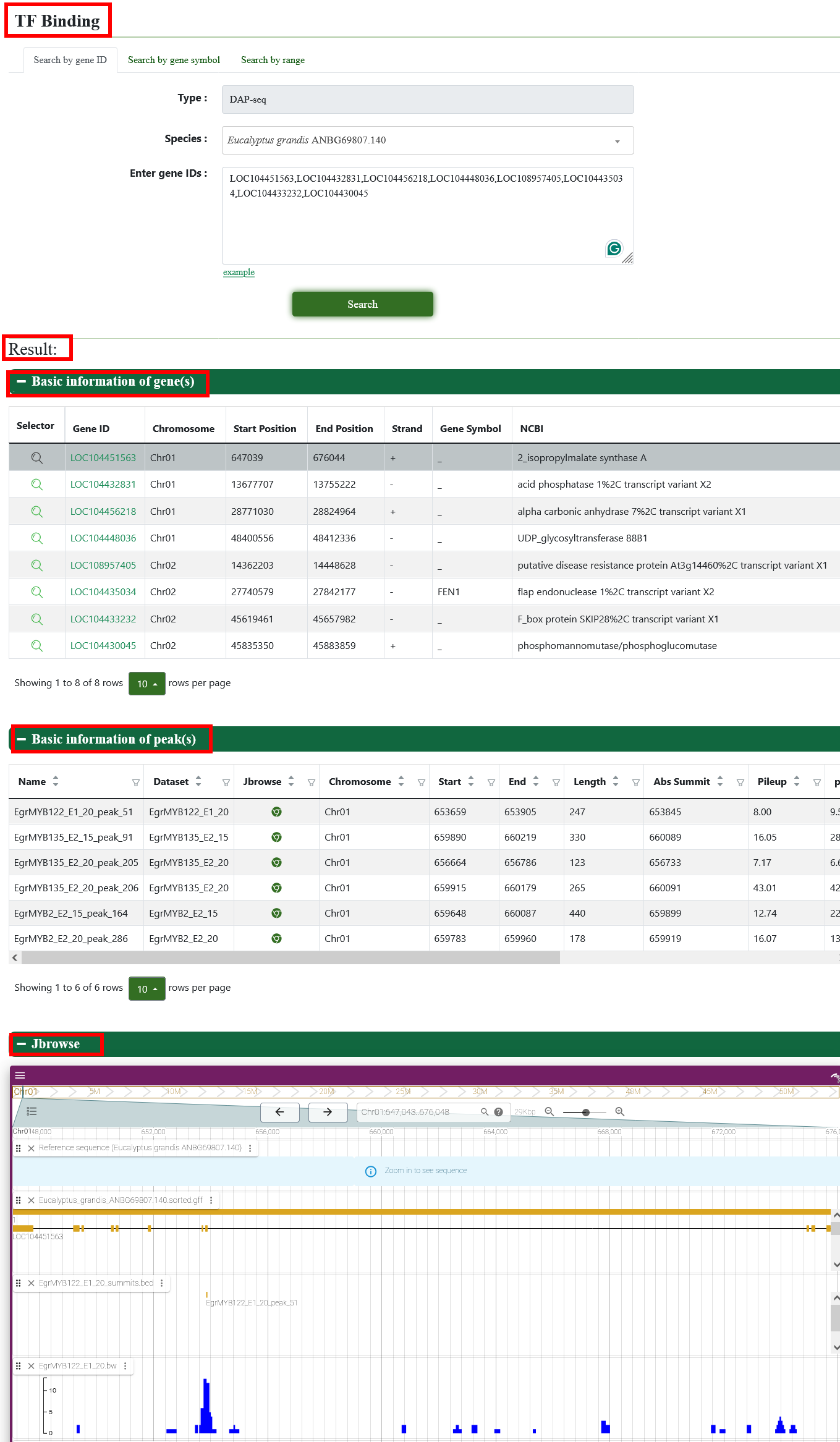
6.2. TF binding
We provide a visual view of transcription factor binding in Eucalyptus grandis on the page and click to Jbrowse page.
7. Variaomics
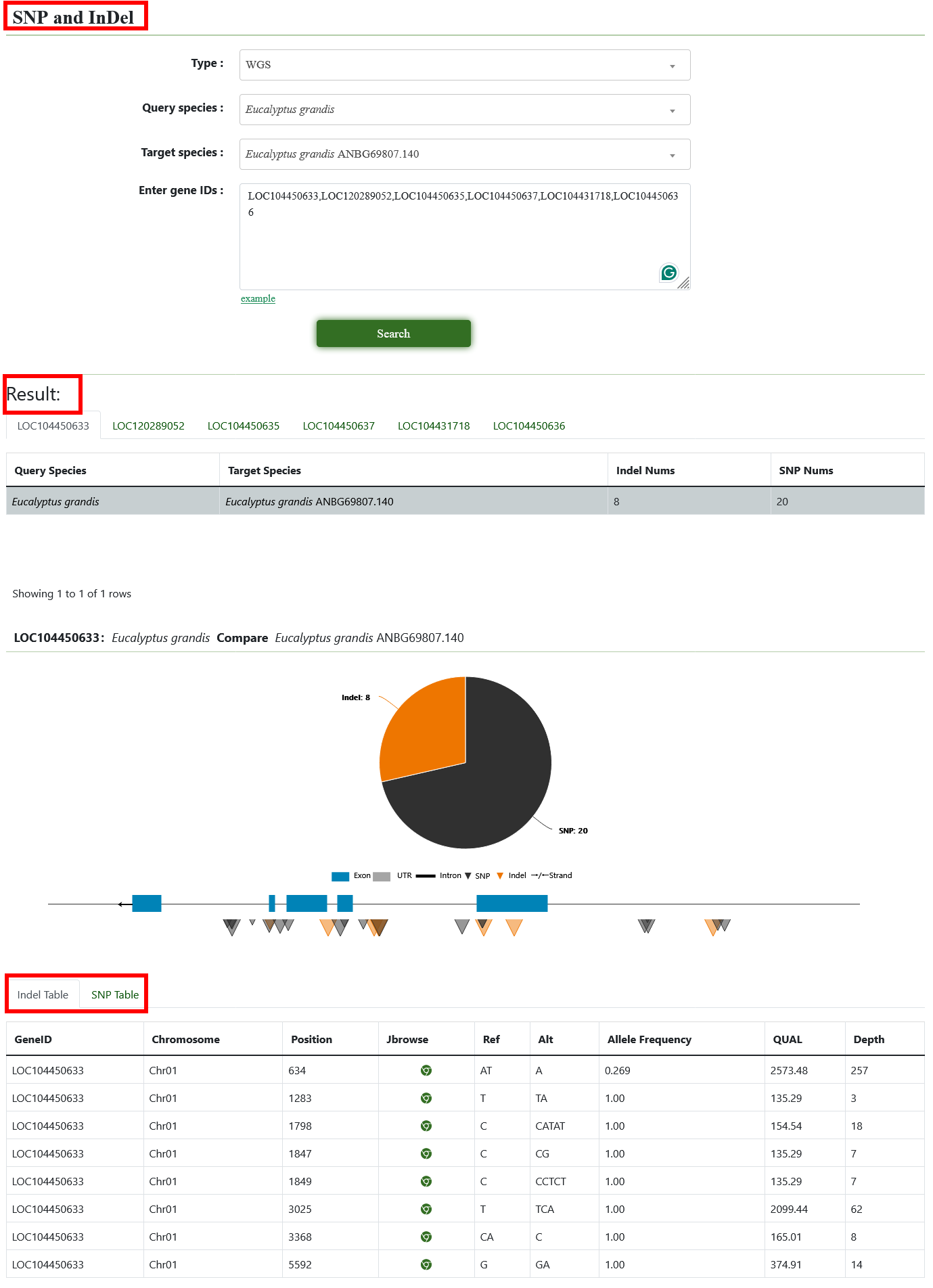
7.1. SNP and InDel
You can obtain the SNP and InDel information of the interested accessions. Users need to select the interested Genomic and the type and choose the Gene ID. After submission, the page shows SNP and InDel SNPs/InDels with the gene structures, and their positions in the upstream and downstream regions, and click to Jbrowse page.
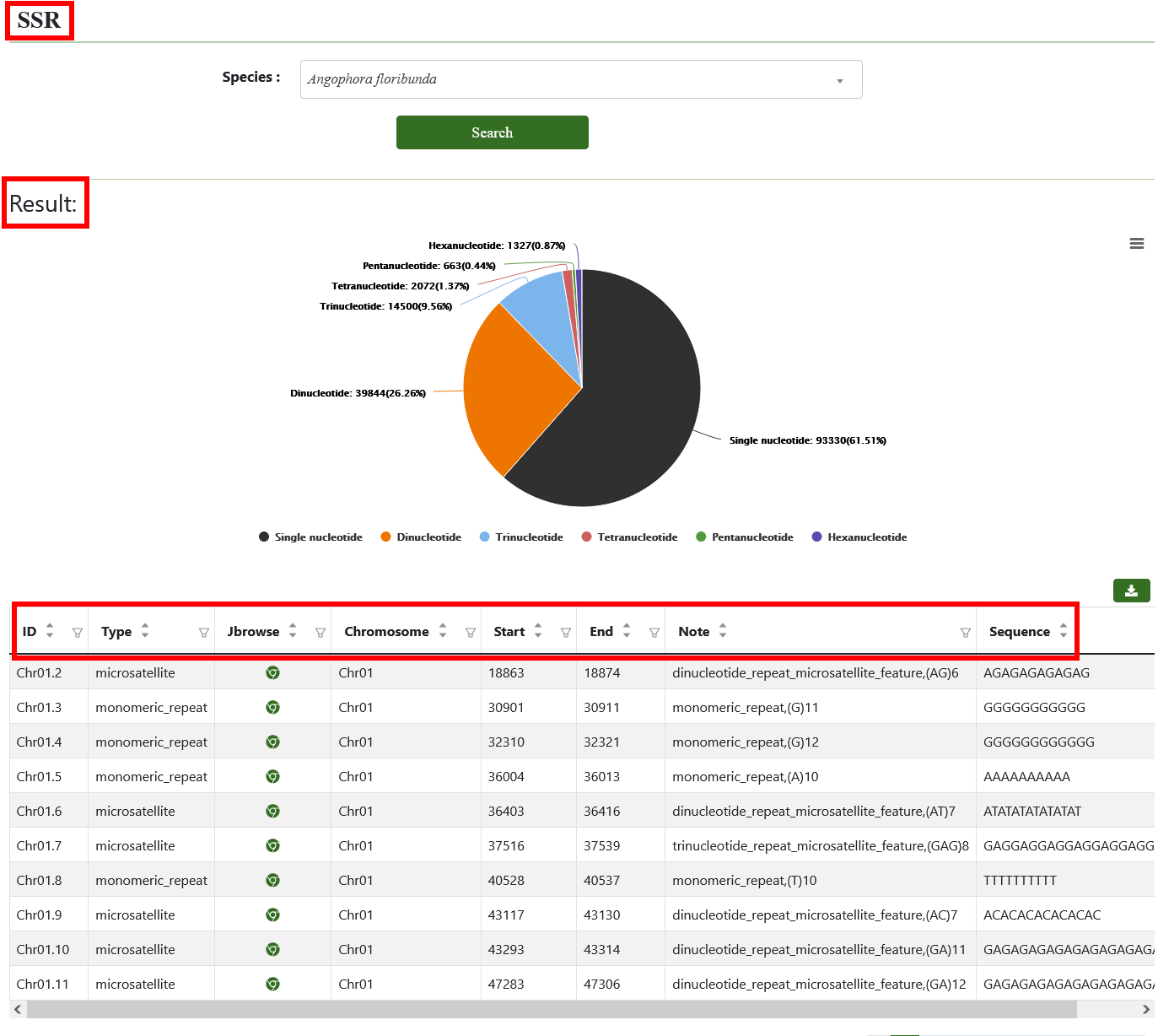
7.2. SSR
We predicted the simple sequence repeats in 45 Eucalyptus genomes using MISA and accessed the results through this page and the JBrowse page.
8. Tools
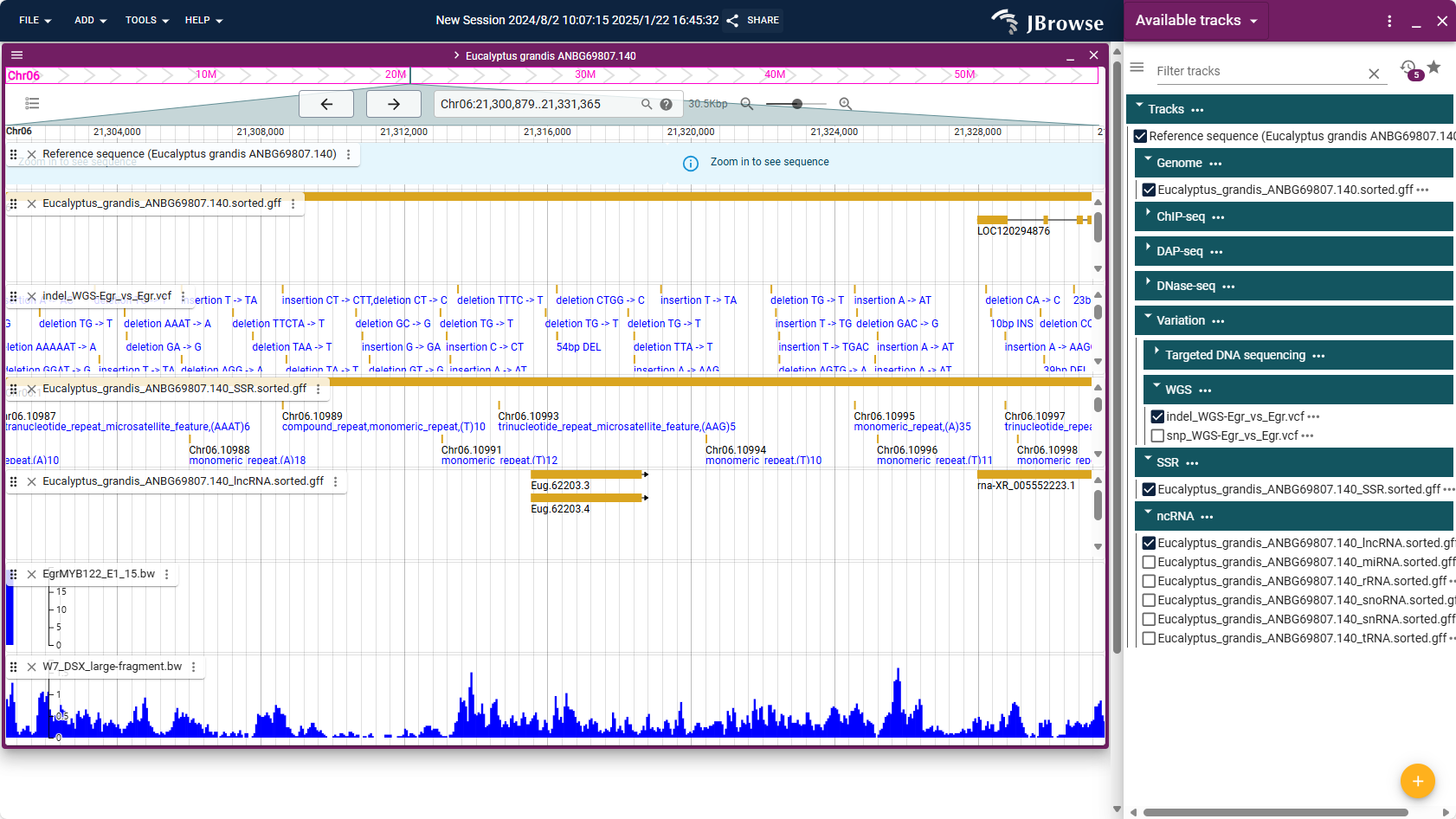
8.1. JBrowse
Users can select different omics data through JBrowse to view all the tracks on the genome, so as to explore the multi-omics data related to the gene of interest.
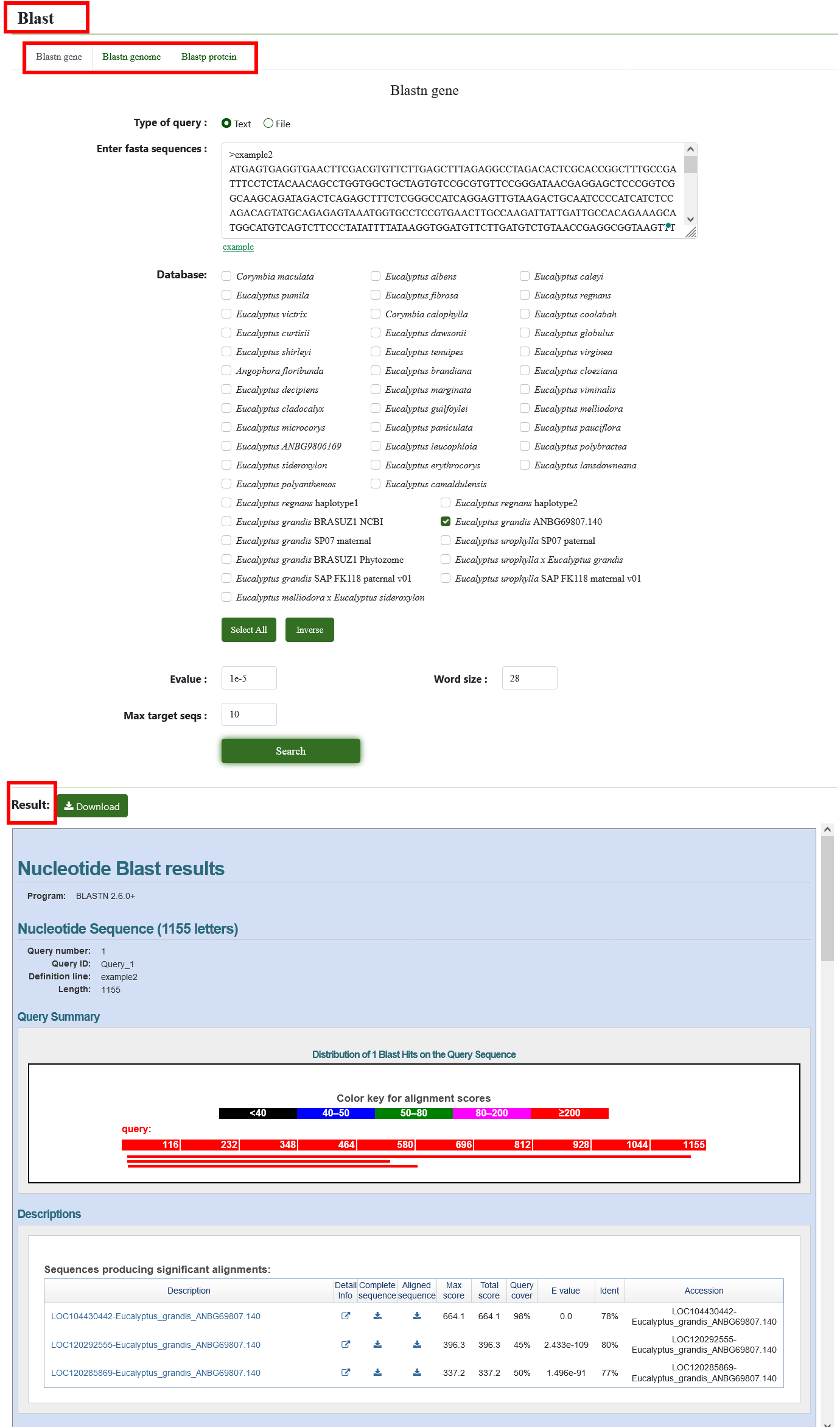
8.2. Blast
You can use BLAST to find homologous sequences of interested genes. Users need to enter the fasta format sequence in the search box or directly drag the file into the search box, select the Blast database to be compared, and select software to use (BLASTN and BLASTP are supported). After submission, we will show the results of the BLAST alignment.
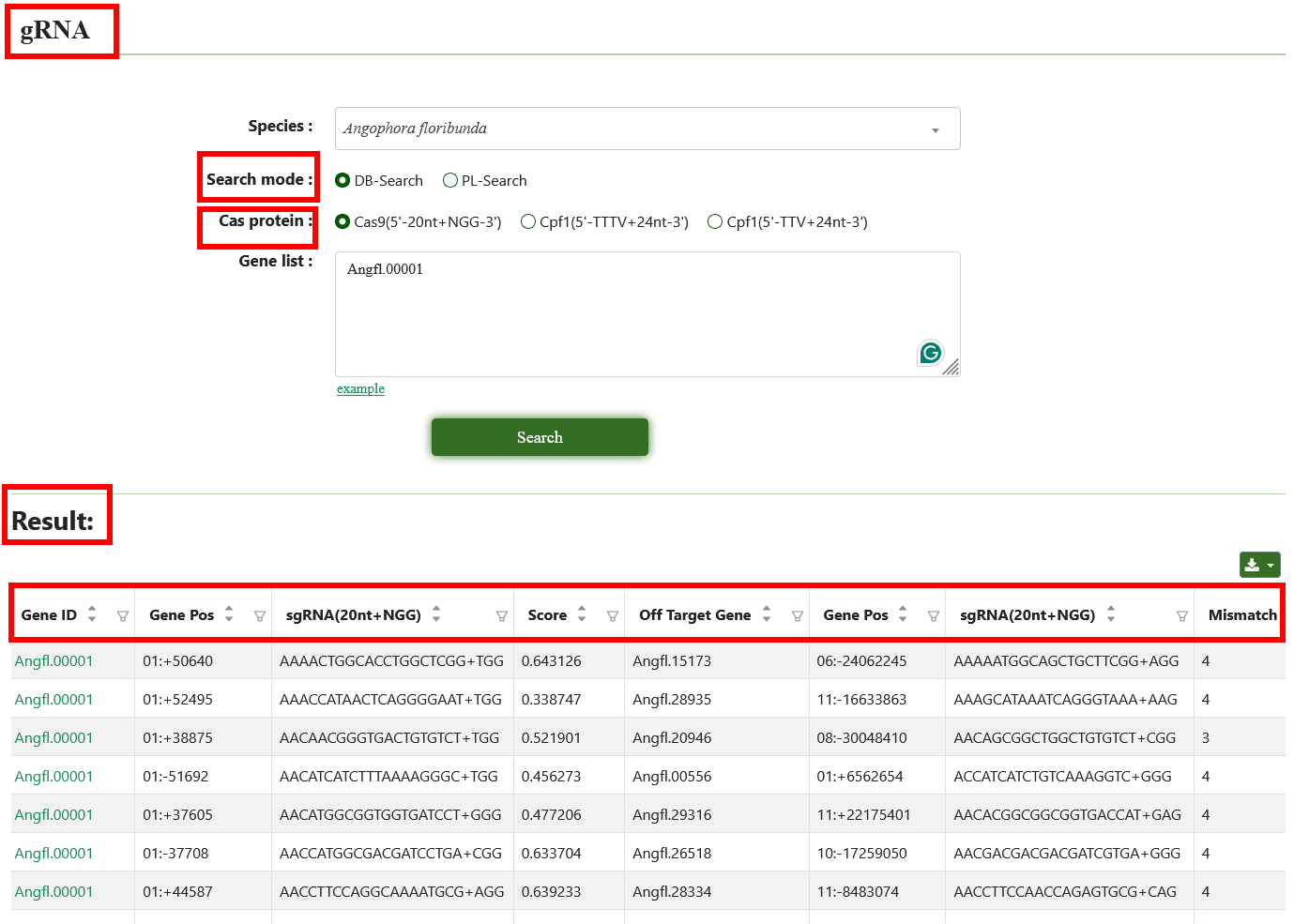
8.3. gRNA
The gRNA tool is an innovative feature designed for selecting guide RNA (gRNA) sequences for gene editing of specific genes across 45 Eucalyptus genomes. To view the results, select a genome, search mode, and Cas protein type, input a gene list, and the possible on-target sgRNAs will be displayed in a table.
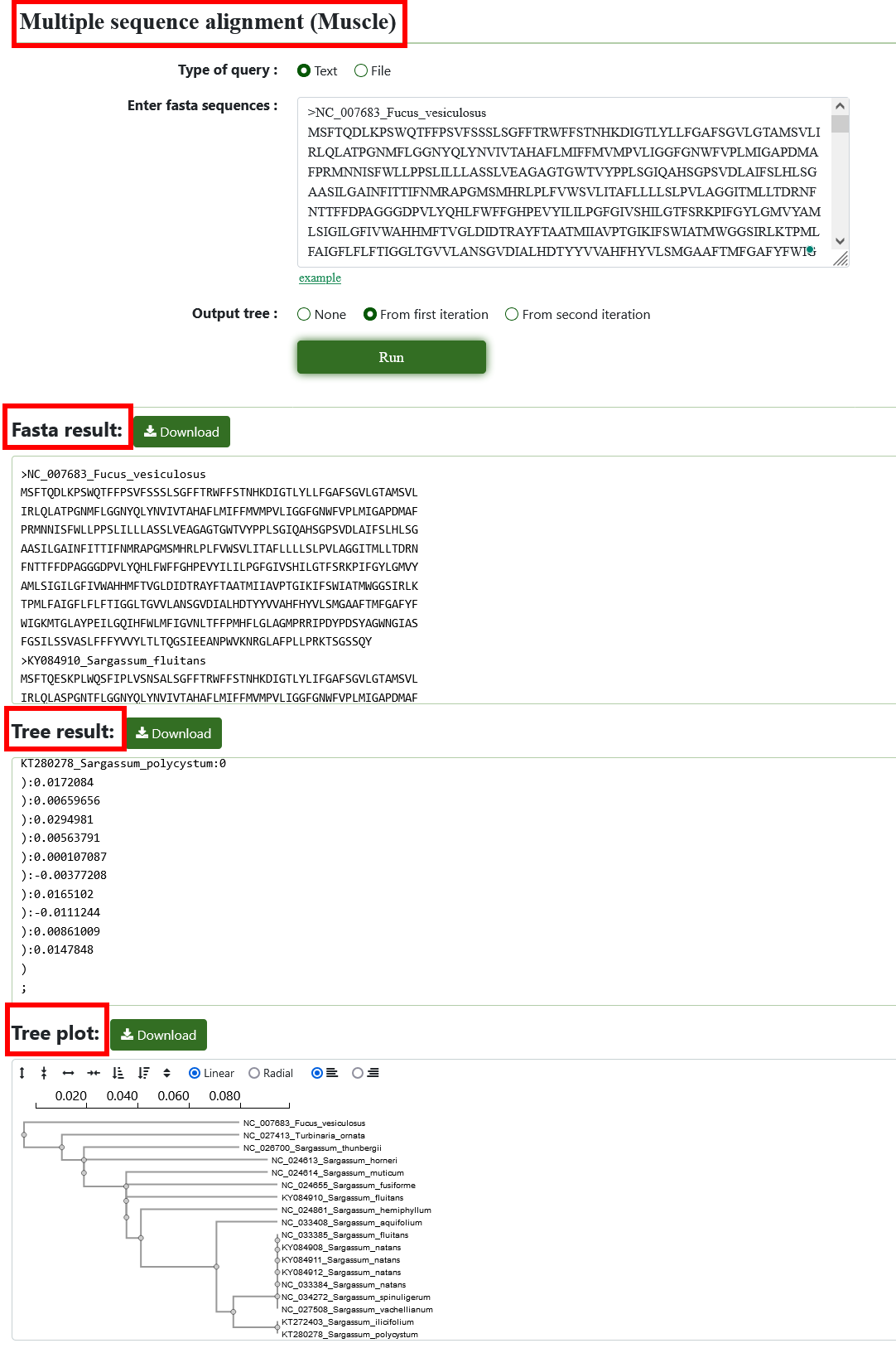
8.4. Multiple sequence alignment (Muscle)
Users can input or upload multiple sequences, choose the style of output tree, and get the results of multi-sequence comparison after submission.
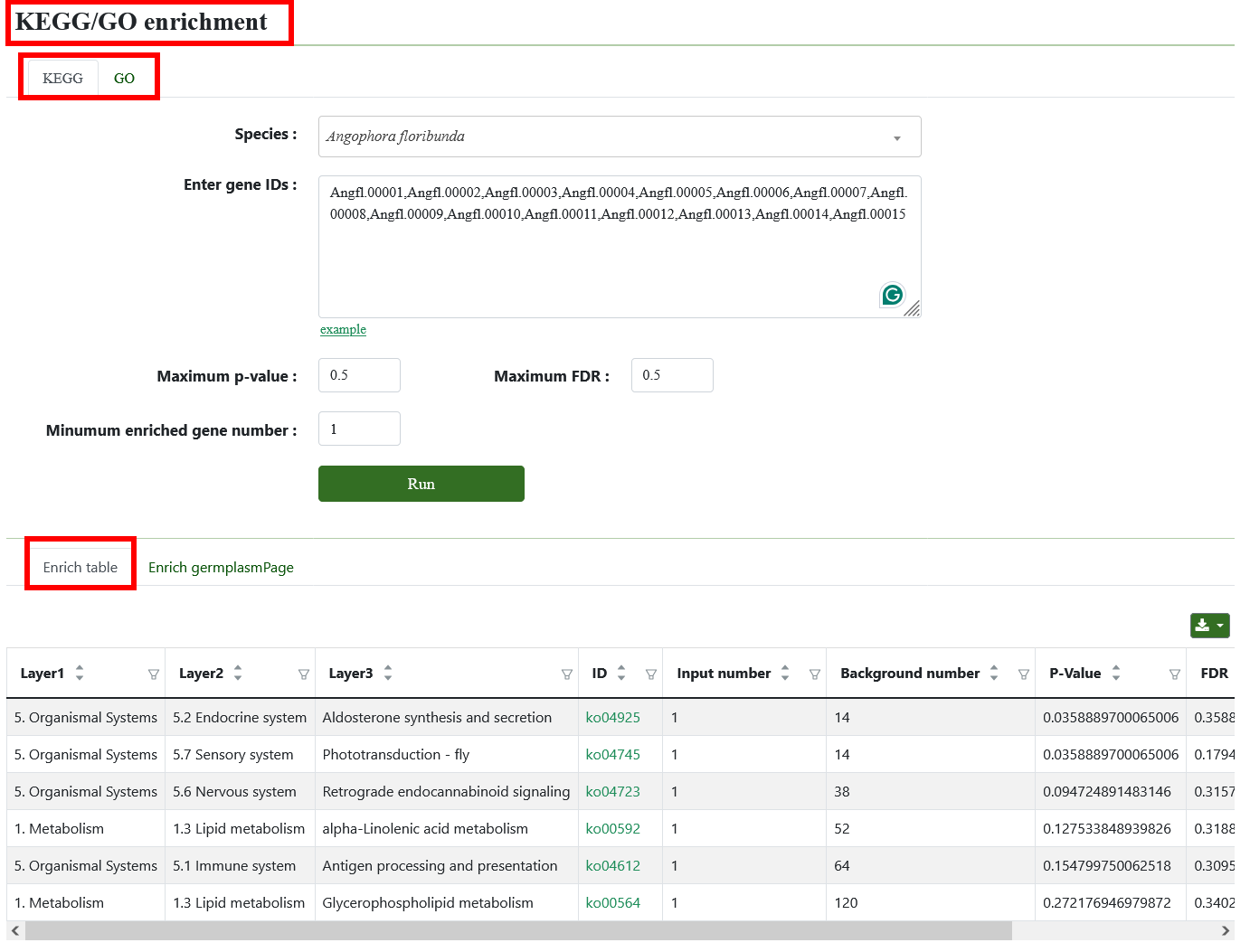
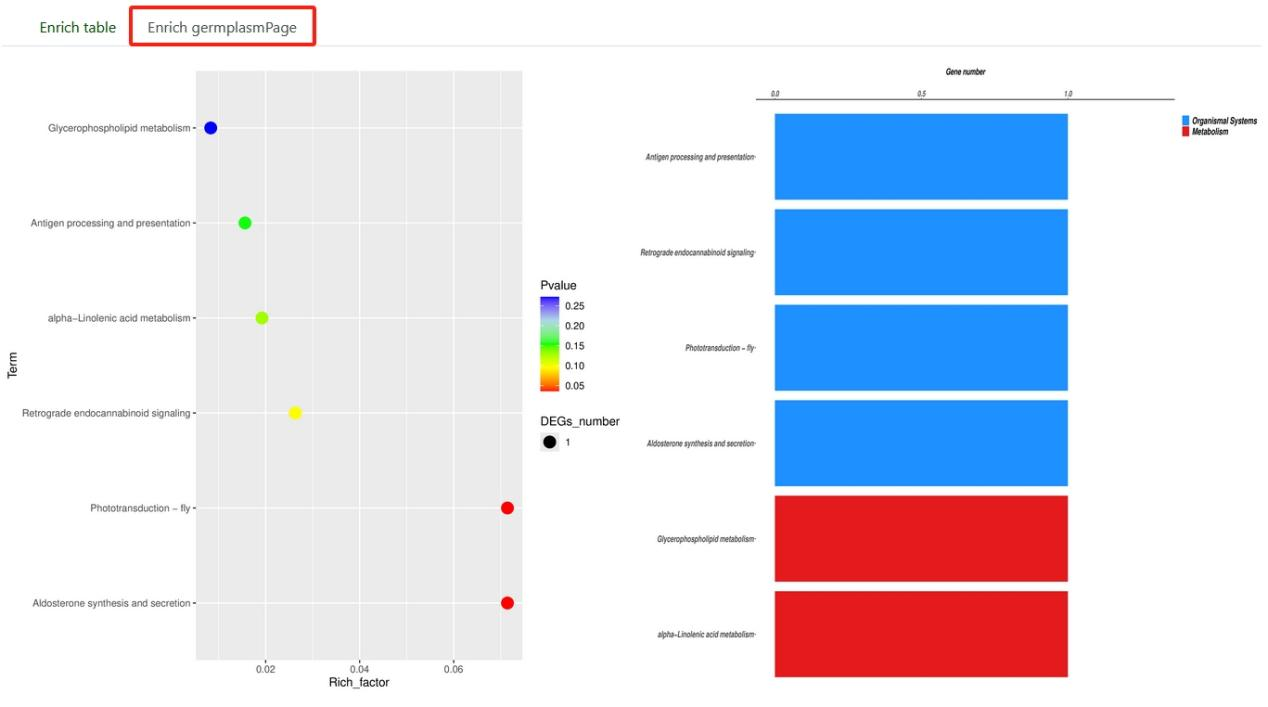
8.5. KEGG/GO enrichment
KEGG/GO enrichment tool is used to evaluate which biological pathways in KEGG/GO database are significantly enriched in gene sets. Users can select the genome and paste the gene list in the sequence box. After submission, we will get enrichment analysis result table, a point chart and a bar chart.
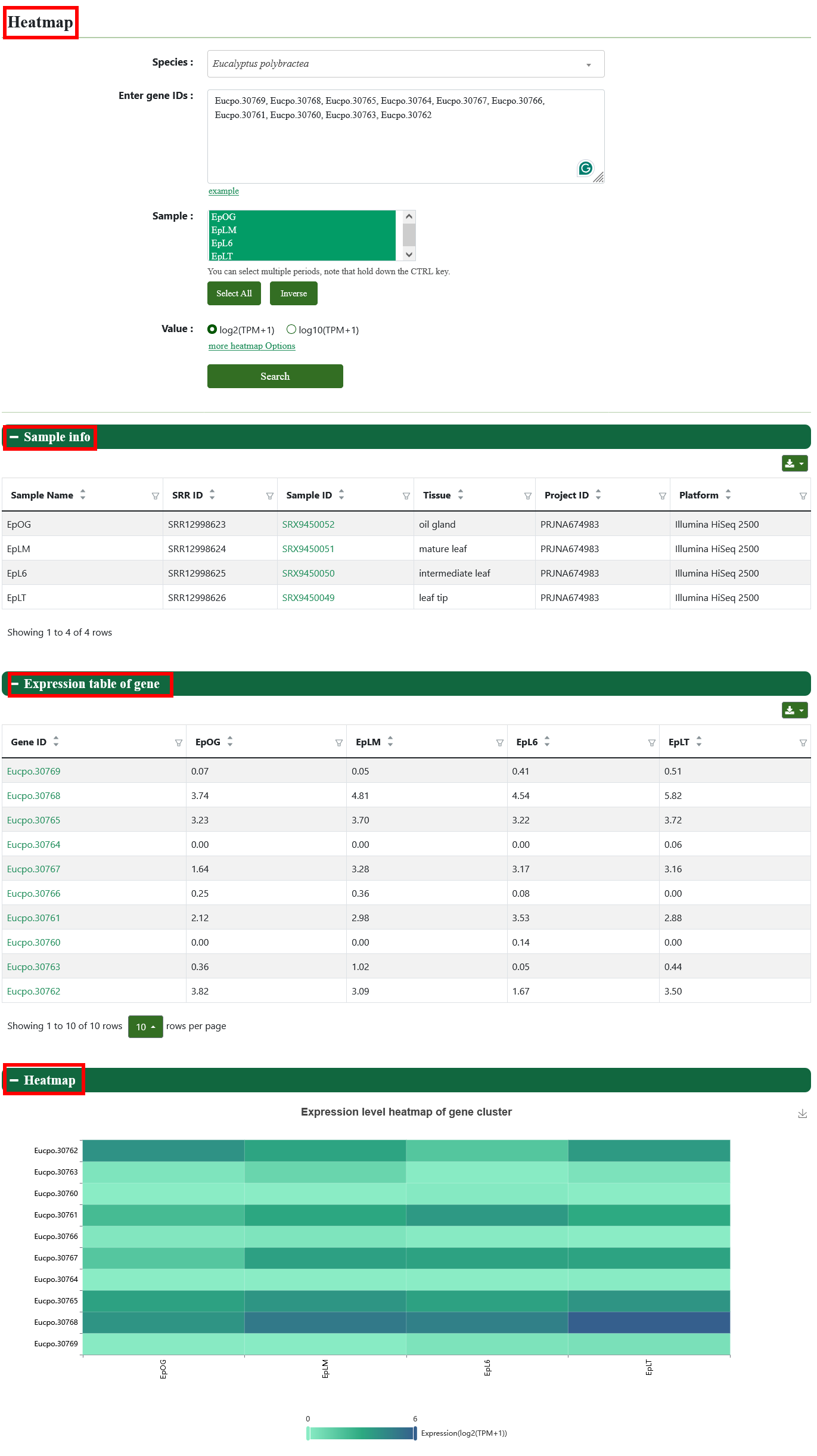
8.6. Heatmap
We visualize the data in the form of heatmap. Users can select genome, genes of interest and transcriptome data, and this page will display the sample information, expression matrix and expression heatmap.
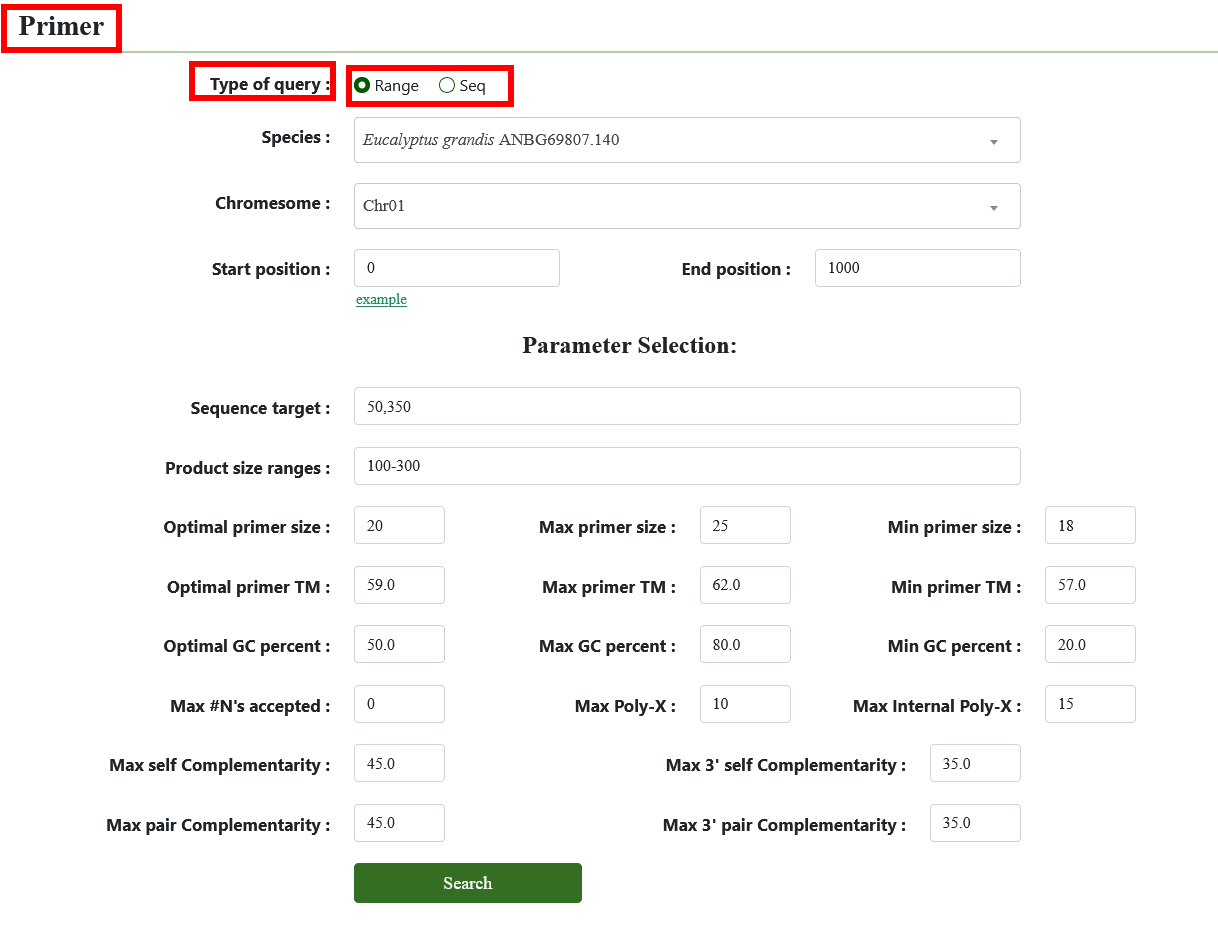
8.7. Primer
Primer3 is for generating primer sequences specific to the target gene. To design primers, adjust the parameters as needed, enter genomic region, and submit; the results will include statistical data for primer design (available for download) and the raw output from Primer3, with visualized primer sequences and their genome positions.
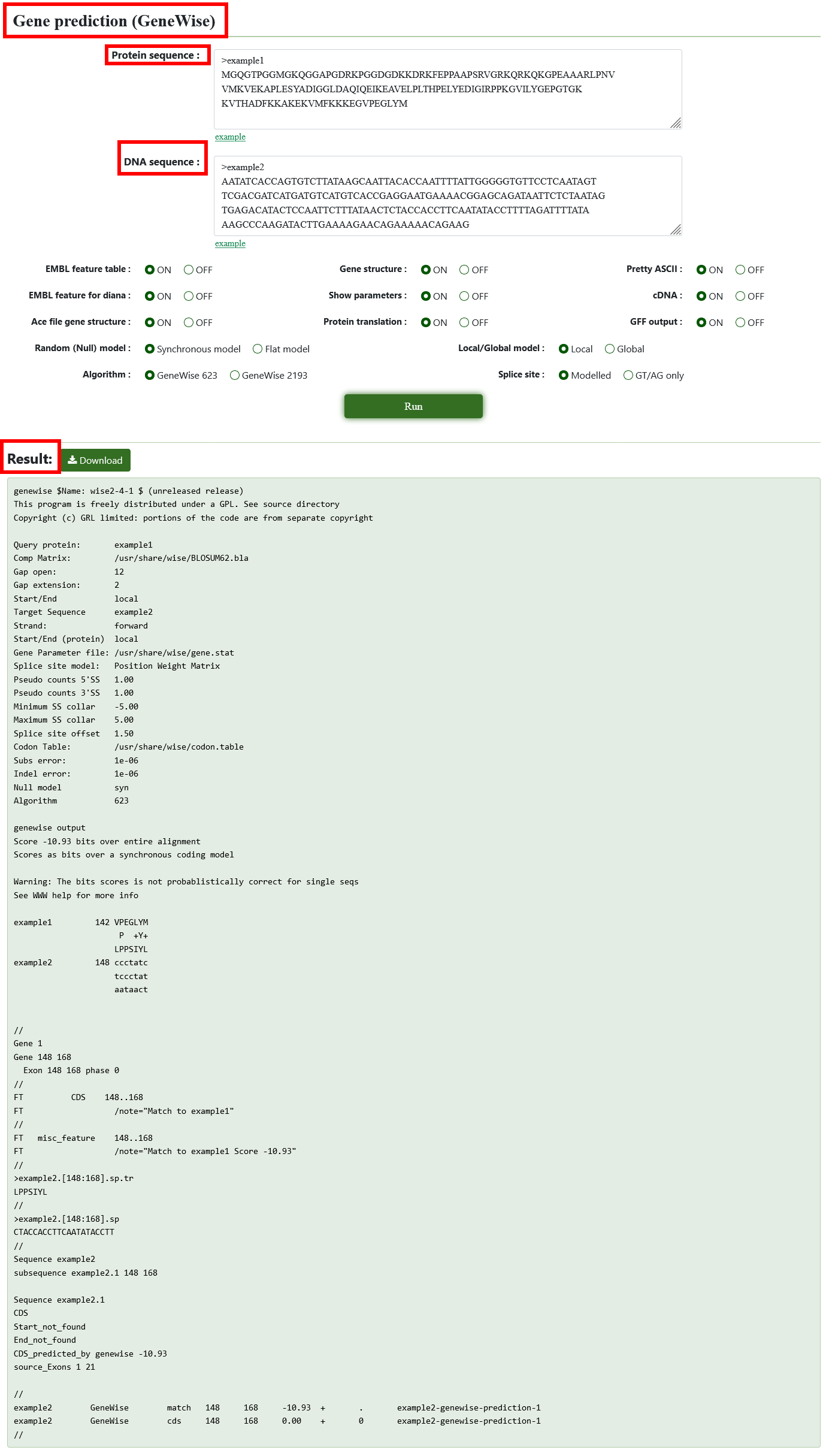
8.8. Gene prediction (GeneWise)
GeneWise is a tool for gene prediction based on homologous proteins. We only need to upload the protein sequence and the target DNA sequence in turn, modify the parameters as needed, and click the "run" button to get the prediction result.
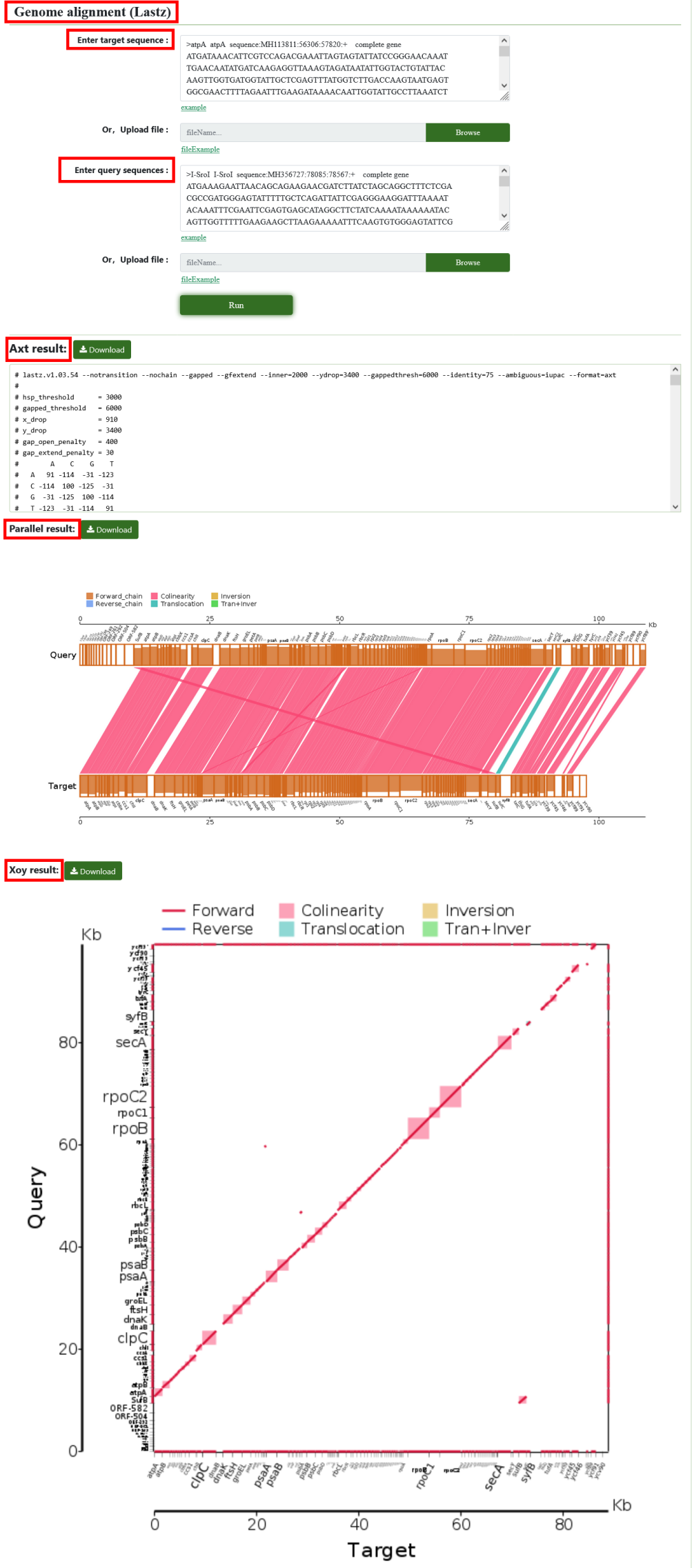
8.9. Genome alignment (Lastz)
LASTZ is a tool for comparing two DNA sequences, which can find similar regions between the two sequences and generate comparison results. Users can submit target sequences and query sequences and get the results.
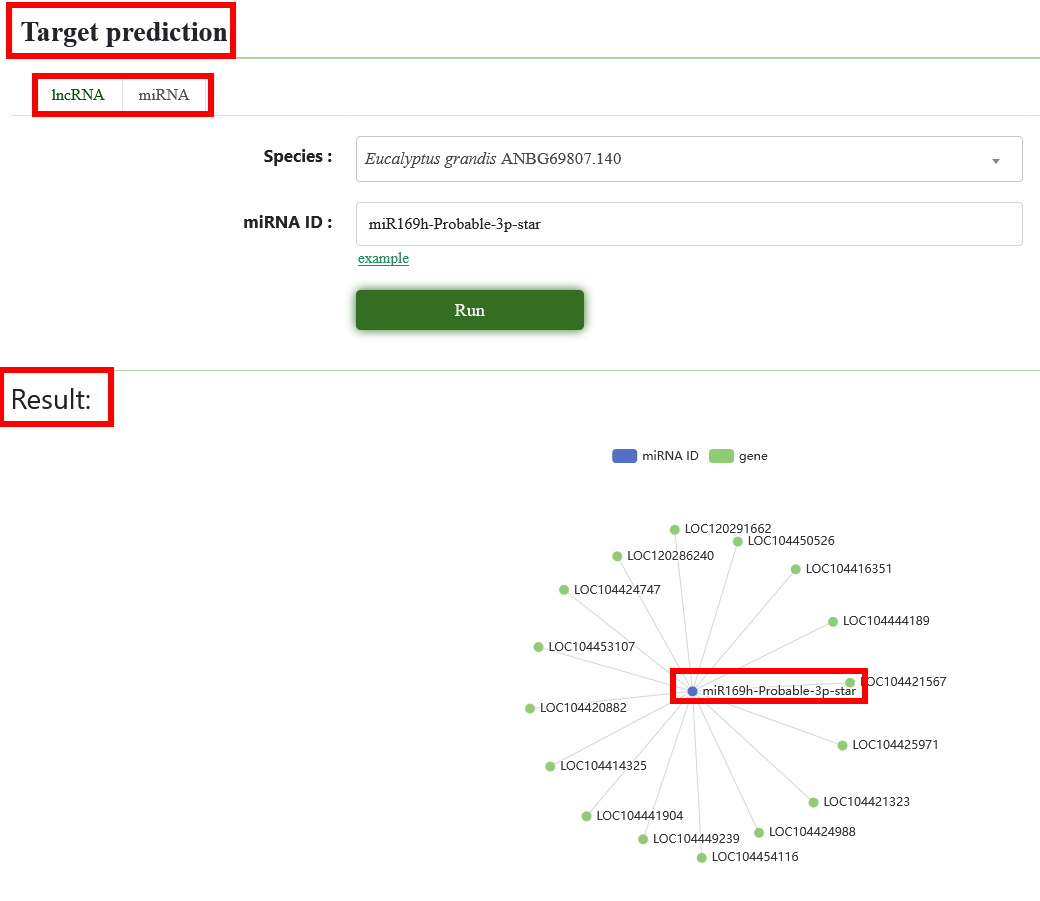
8.10. Target prediction
We predict the miRNA and lncRNA target sites of the genome. Users need to select the genome of interest, enter the ID, and we will show the network diagram of miRNA/lncRNA and its target genes.
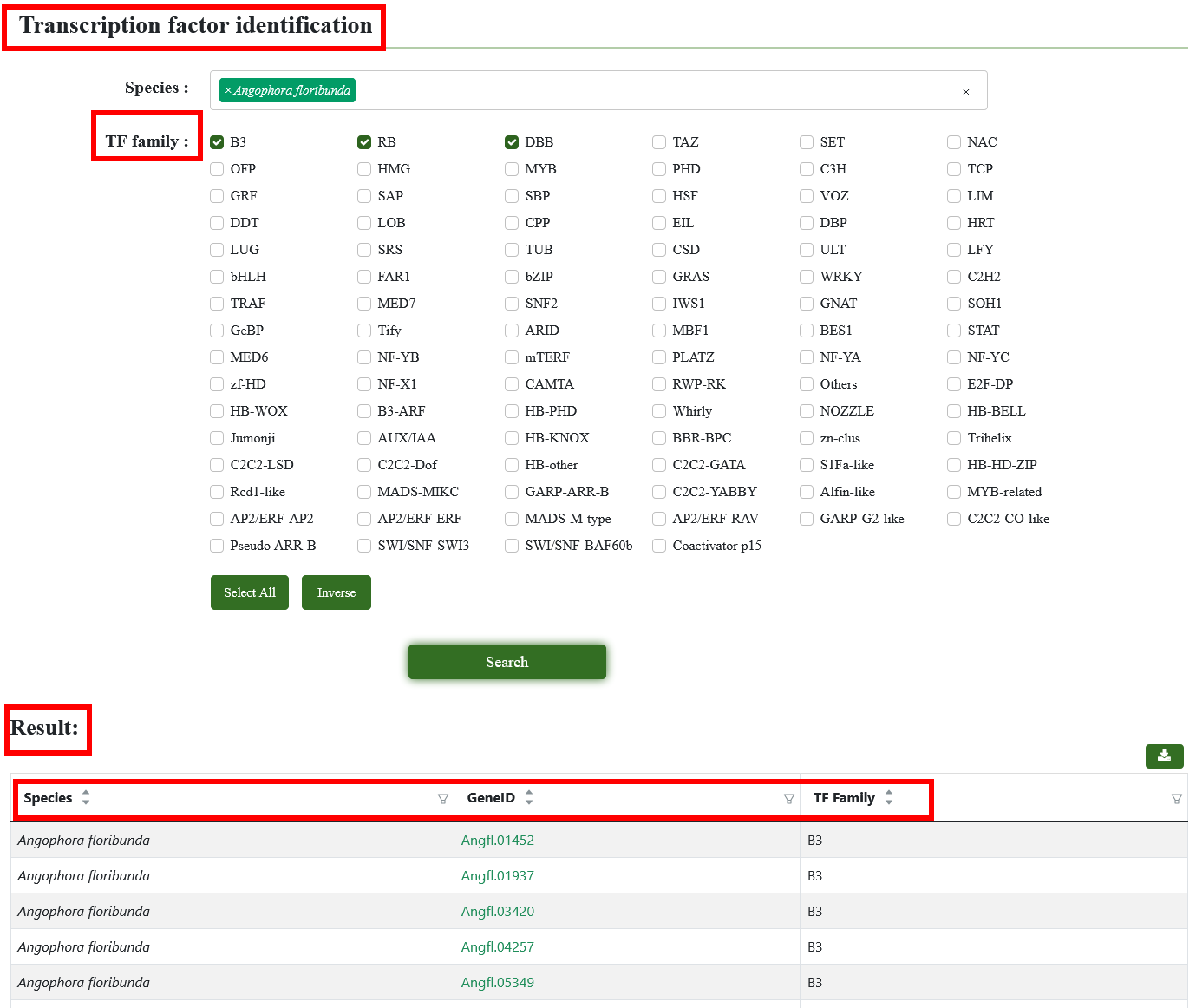
8.11. Transcription factor identification
We provide 94 Transcription factors, users can select one or more TFs, and the interested genome (multiple selections are supported). After submission, we will show a form, including specie, gene ID, TF Name . This form supports retrieval by gene ID and provides functions such as downloading. Click on the Gene ID to jump to the gene detail page.
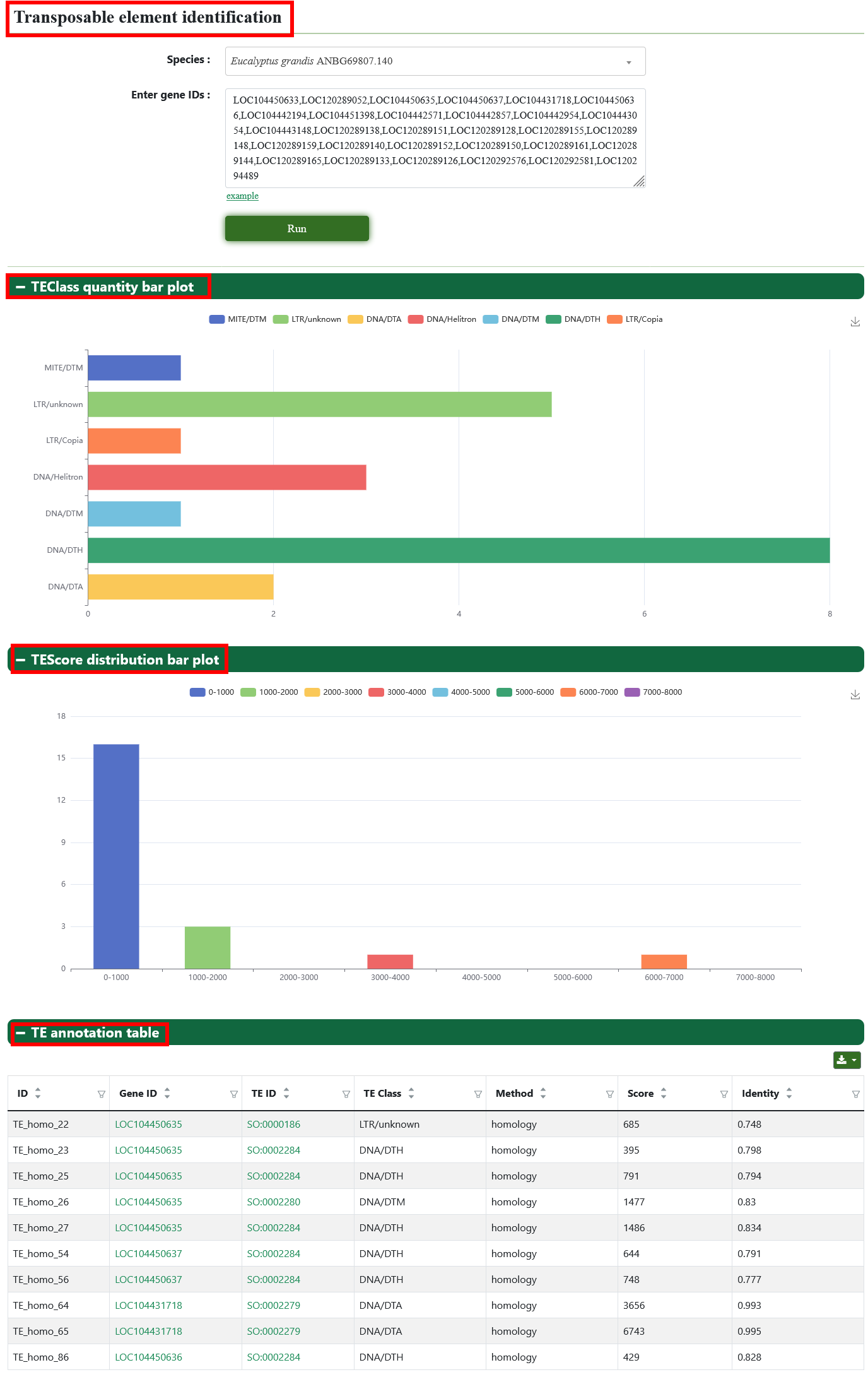
8.12. Transposable element identification
Users can select genome and submit gene ID on this page to get the result of Transposable element identification.
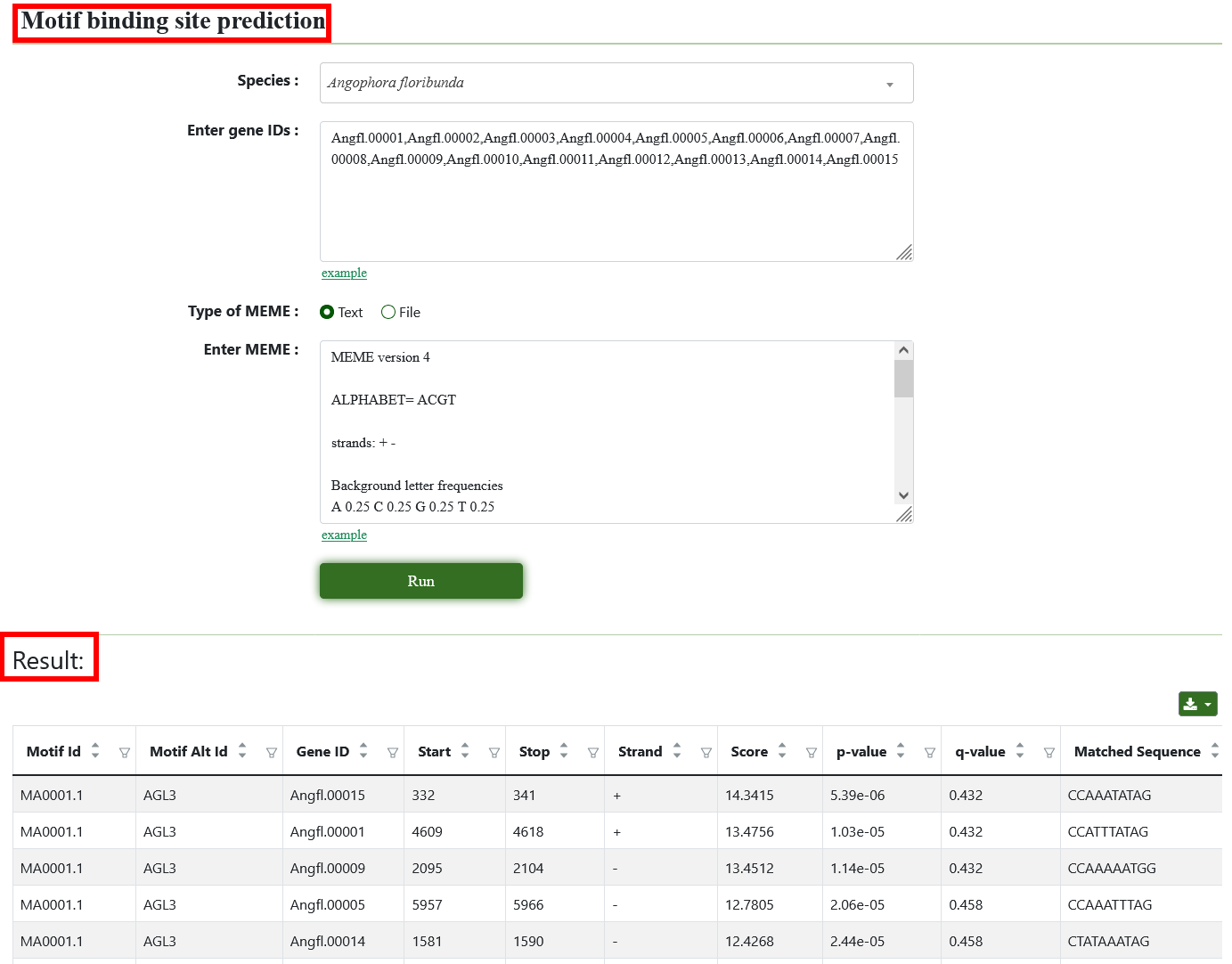
8.13. Motif binding site prediction
The tool is to search for known motif in a given DNA sequence. Users need to provide gene ID and Motif databases, and then locate the positions of these motifs in the sequence and output information such as the position and score of each motif.
9. Download
Users can download genome sequences, annotation files, CDS, protein, and transcript sequences, as well as the sample information table from the Download module.
10. Help
The Help module provides the user manual and the author's information.