Welcome to EucaMOD
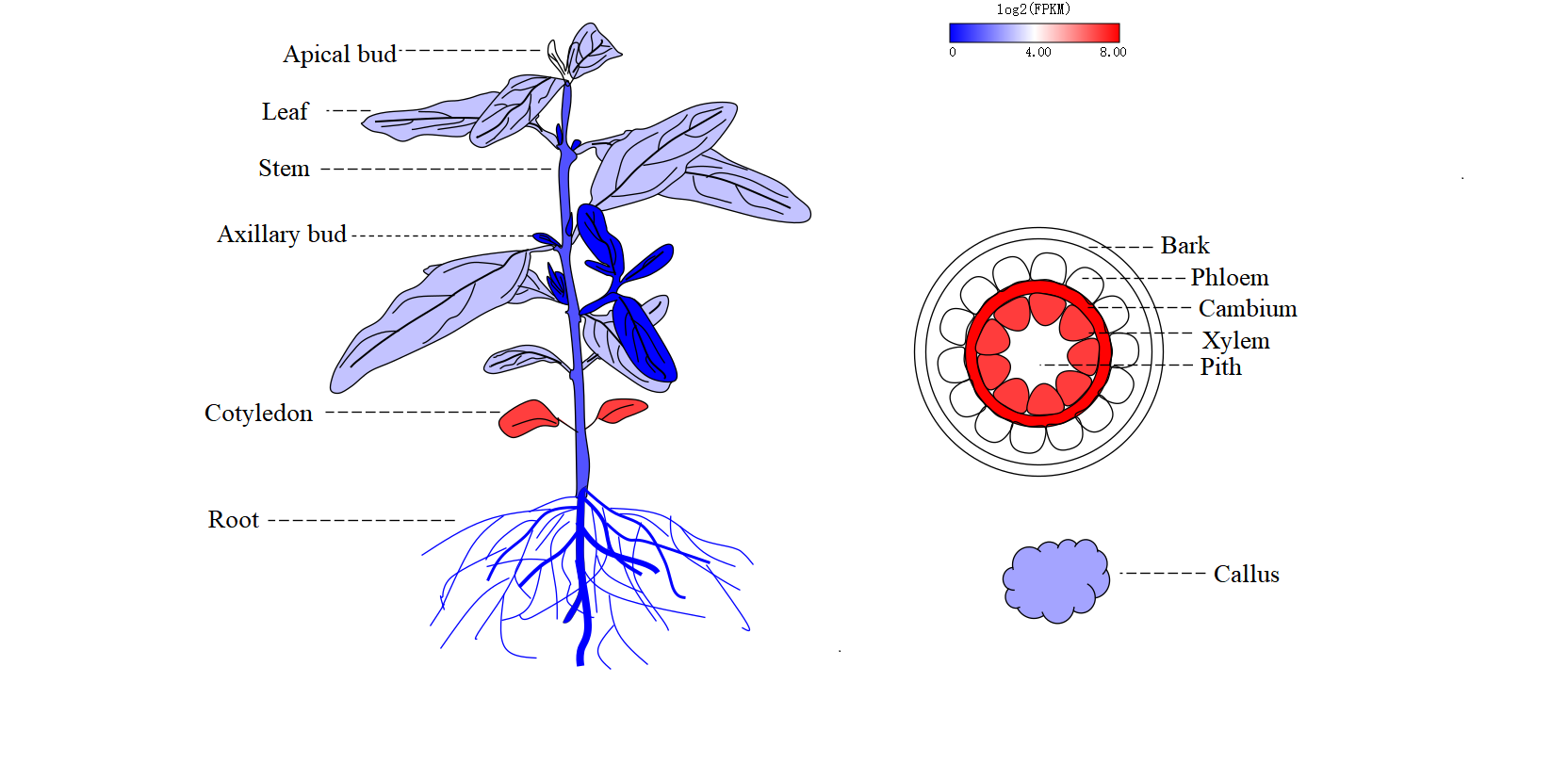
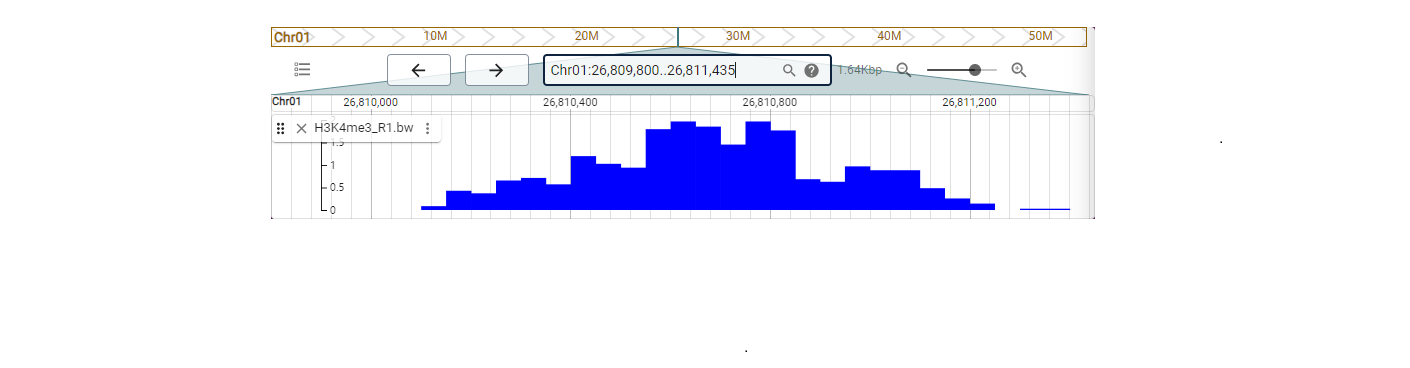
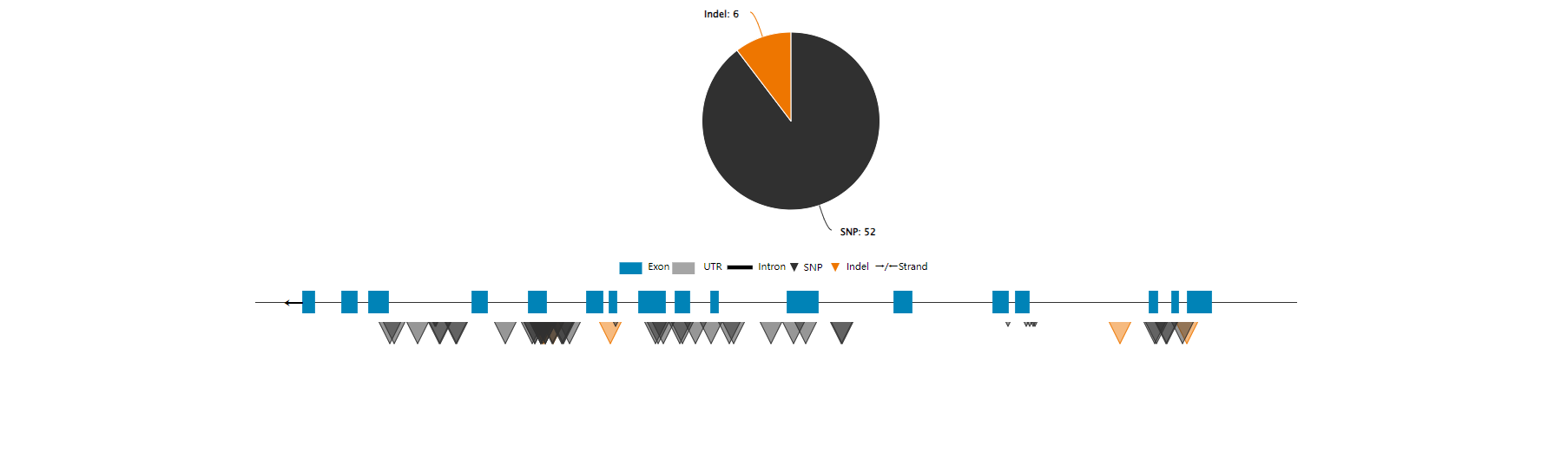
Eucalyptus multi-omics database (EucaMOD) integrates a wide range of omics data, including genomics, comparative genomics, pan-proteomics, transcriptomics, epigenetics, and variomics. It also features 13 powerful analysis tools, such as JBrowse, Blast, gRNA, multiple sequence alignment (Muscle), KEGG/GO enrichment, heatmap, primer design, gene prediction (GeneWise), genome alignment (Lastz), target prediction of lncRNAs and miRNAs, transcription factor identification, transposable element identification, and motif binding site prediction. By incorporating JBrowse, EucaMOD facilitates seamless browsing of multi-omics data and significantly simplifies in-depth analysis and interpretation of Eucalyptus multi-omics data in an intuitive manner.
See Tutorial page for more help.
Data Statistics
2207
Samples
45
Genomes
39
Species
13
Tools
Loading, please wait
Type |
Number of samples |
Number of species |
Characteristic |
|---|---|---|---|
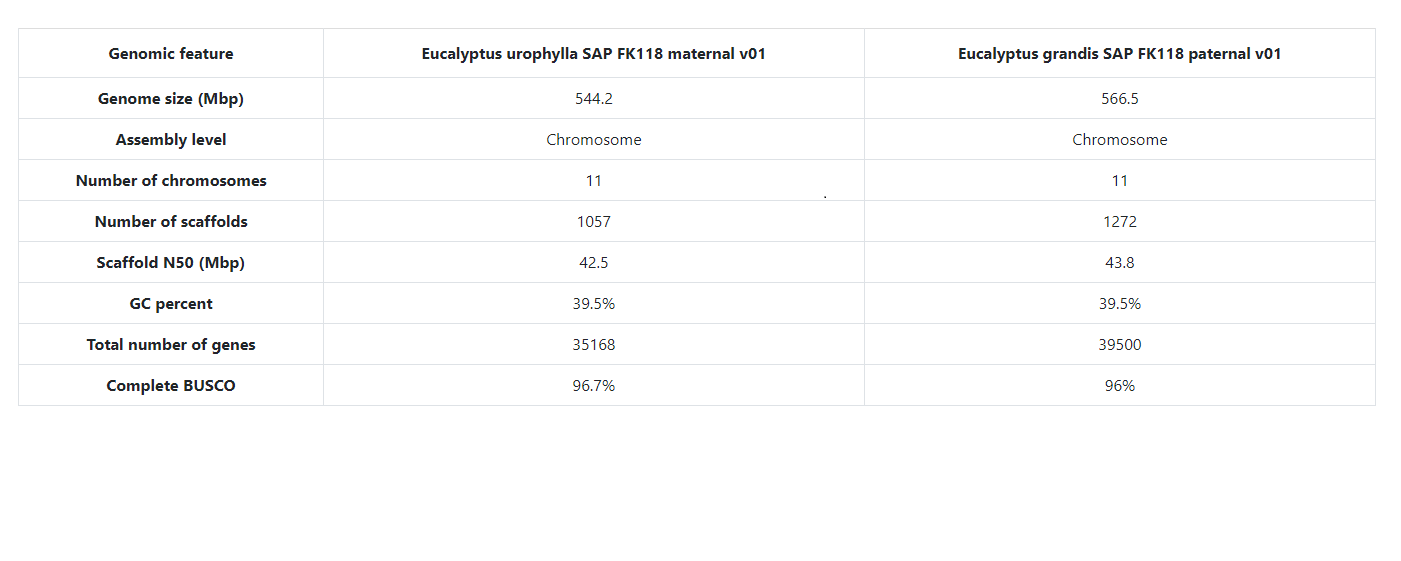
| Genome Assembly | 45 | 39 | 2,064,765 genes |
| RNA-seq | 870 | 10 | 703,774 transcripts |
| miRNA-seq | 17 | 3 | 797 miRNAs |
| SNP variation | 1,219 | 11 | 46,310,119 variations |
| Indel variation | 1,219 | 11 | 8,398,921variations |
| DAP-seq | 26 | 1 | 33,432 peaks |
| DNase-seq | 12 | 1 | 712,334 peaks |
| ChIP-seq | 14 | 1 | 234,680 peaks |